Exploring the Power of Python Libraries: A Deep Dive into Pandas
Table of Contents
ToggleWhat is Pandas?
Pandas is a Python library used for working with data sets.
It includes functions for analyzing, cleaning, examining, and modifying data.
The word “Pandas” refers to both “Panel Data” and “Python Data Analysis” and was created by Wes McKinney in 2008.
Importing Pandas
- You should be able to import Pandas after installing it
- We’ll import pandas as its alias name pd
import pandas as pd
import numpy as np
Introduction: Why to use Pandas?
How is it different from numpy ?
- The major limitation of numpy is that it can only work with 1 datatype at a time
- Most real-world datasets contain a mixture of different datatypes
- Like names of places would be string but their population would be int
==> It is difficult to work with data having heterogeneous values using Numpy
Pandas can work with numbers and strings together
So lets see how we can use pandas
Reading dataset in Pandas
df= pd.read_csv(r"C:\Users\welcome\Downloads\dataset1.csv")
Now how should we read this dataset?
Pandas makes it very easy to work with these kinds of files
df
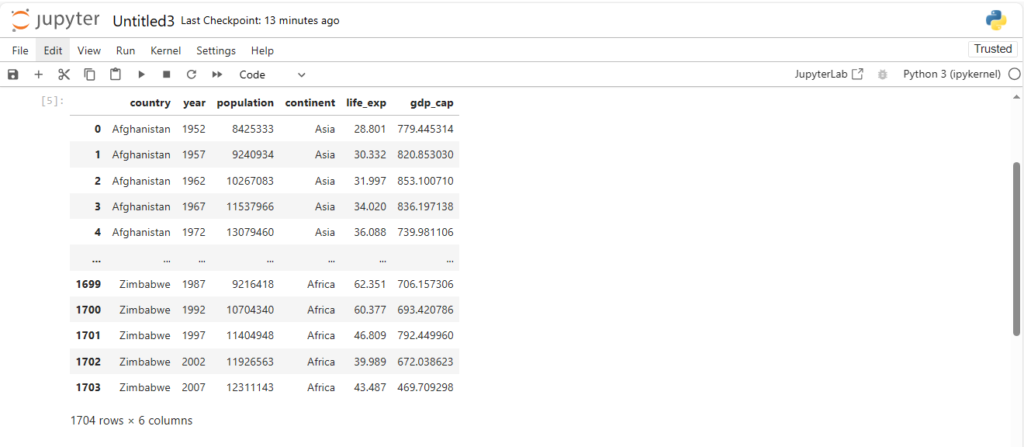
Dataframe and Series
What can we observe from the above dataset ?
We can see that it has:
- 6 columns
- 1704 rows
what do you think is the datatype of df ?
type(df)
pandas.core.frame.DataFrame
What is a pandas DataFrame ?
- It is a table-like representation of data in Pandas => Structured Data
- Structured Data here can be thought of as tabular data in a proper order
- Considered as counterpart of 2D-Matrix in Numpy
Now how can we access a column, say country of the dataframe?
df["country"]
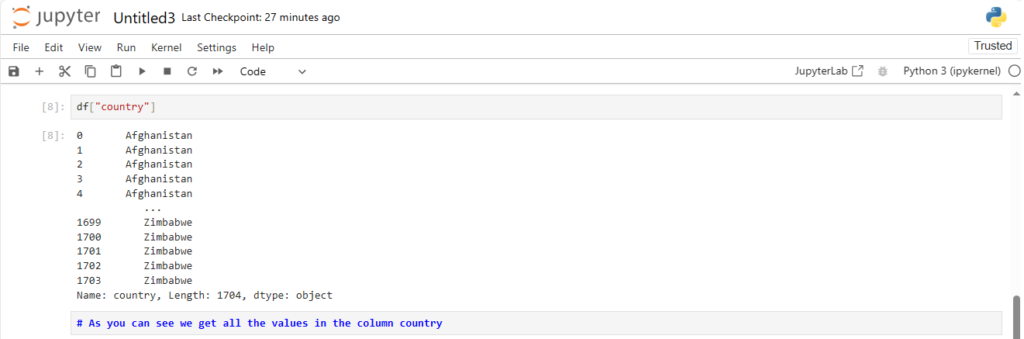
Now what is the data-type of a column?
type(df["country"])
pandas.core.series.Series
Its a pandas Series
What is a pandas Series ?
- Series in Pandas is what a Vector is in Numpy
What exactly does that mean?
- It means a Series is a single column of data
- Multiple Series stack together to form a DataFrame
Now we have understood what Series and DataFrames are
What if a dataset has 100 rows … Or 100 columns ?
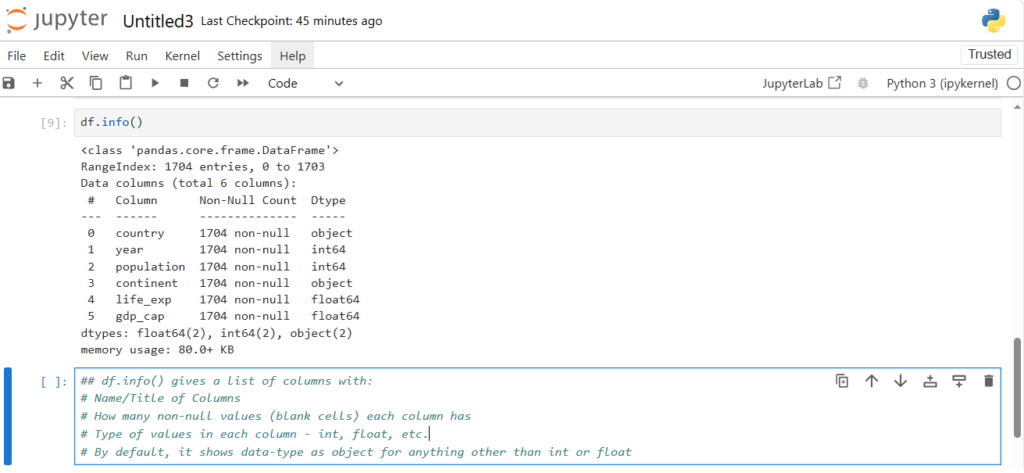
How can we find the datatype, name, total entries in each column ?
df.info()
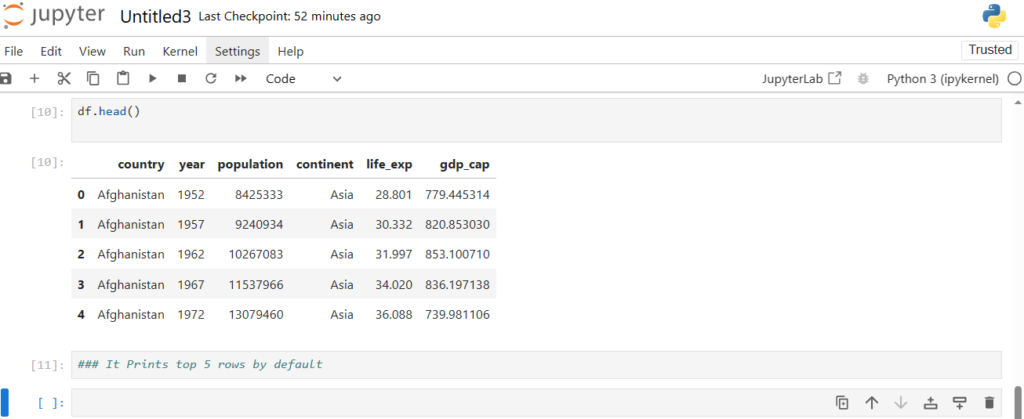
Now what if we want to see the first few rows in the dataset ?
df.head()
We can also pass in number of rows we want to see in head()
df.head(20)
Similarly what if we want to see the last 20 rows ?
df.tail(20) #Similar to head
How can we find the shape of the dataframe?
df.shape
(1704, 6)
Similar to Numpy, it gives No. of Rows and Columns — Dimensions
Now we know how to do some basic operations on dataframes
But what if we aren’t loading a dataset, but want to create our own.
Let’s take a subset of the original dataset
df.head(3) # We take the first 3 rows to create our dataframe
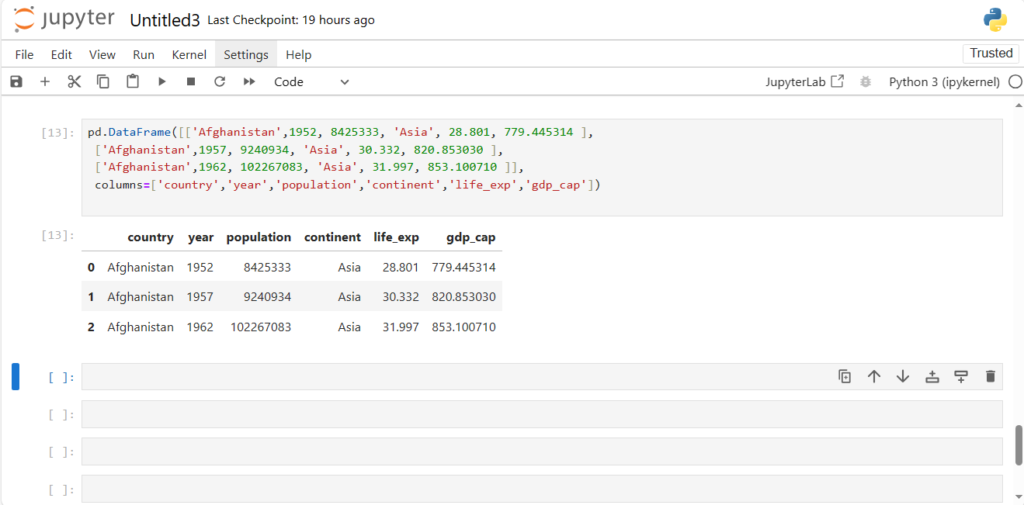
How can we create a DataFrame from scratch?
Approach 1: Row-oriented
- It takes 2 arguments – Because DataFrame is 2-dimensional
- A list of rows
- Each row is packed in a list []
- All rows are packed in an outside list [[]] – To pass a list of rows
- A list of column names/labels
pd.DataFrame([['Afghanistan',1952, 8425333, 'Asia', 28.801, 779.445314 ],
['Afghanistan',1957, 9240934, 'Asia', 30.332, 820.853030 ],
['Afghanistan',1962, 102267083, 'Asia', 31.997, 853.100710 ]],
columns=['country','year','population','continent','life_exp','gdp_cap'])
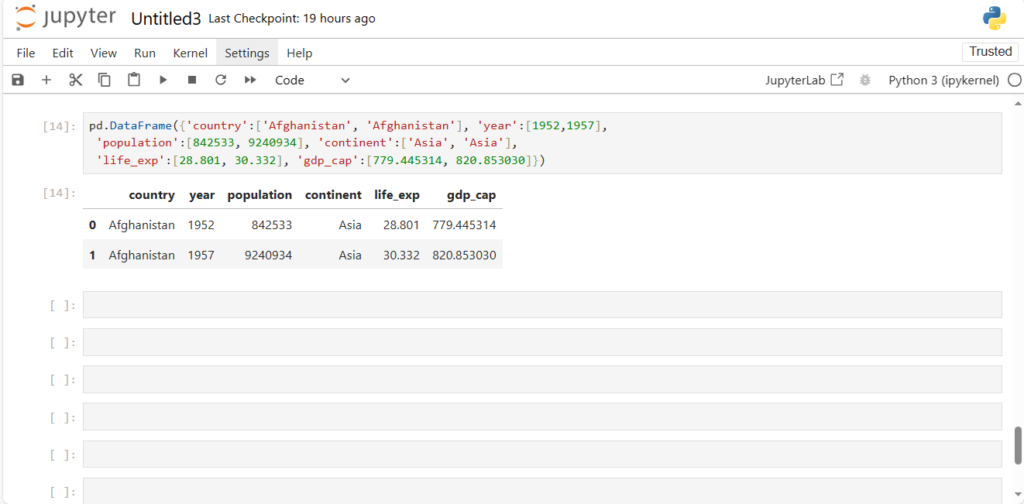
Approach 2: Column-oriented
pd.DataFrame({'country':['Afghanistan', 'Afghanistan'], 'year':[1952,1957],
'population':[842533, 9240934], 'continent':['Asia', 'Asia'],
'life_exp':[28.801, 30.332], 'gdp_cap':[779.445314, 820.853030]}) We pass the data as a dictionary
- Key is the Column Name/Label
- Value is the list of values column-wise
- We now have a basic idea about the dataset and creating rows and columns
What kind of other operations can we perform on the dataframe?
Thinking from database perspective: - Adding data
- Removing data
- Updating/Modifying data
and so on
How can we get the names of all these cols ?
We can do it in two ways:
1. df.columns
2. df.keys
df.columns # using attribute `columns` of dataframe
Index(['country', 'year', 'population', 'continent', 'life_exp', 'gdp_cap'], dtype='obje
ct')
df.keys() # using method keys() of dataframe
Index(['country', 'year', 'population', 'continent', 'life_exp', 'gdp_cap'], dtype='obje
ct')
Note:
- Here, Index is a type of pandas class used to store the address of the series/dataframe
- It is an Immutable sequence used for indexing and alignment.
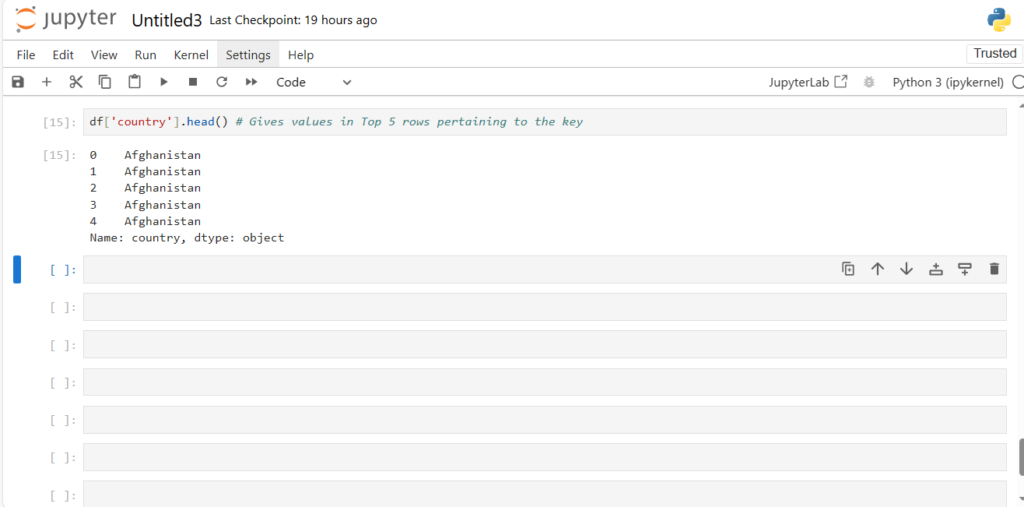
df['country'].head() # Gives values in Top 5 rows pertaining to the key
But what is so “special” about this dictionary?
It can take multiple keys
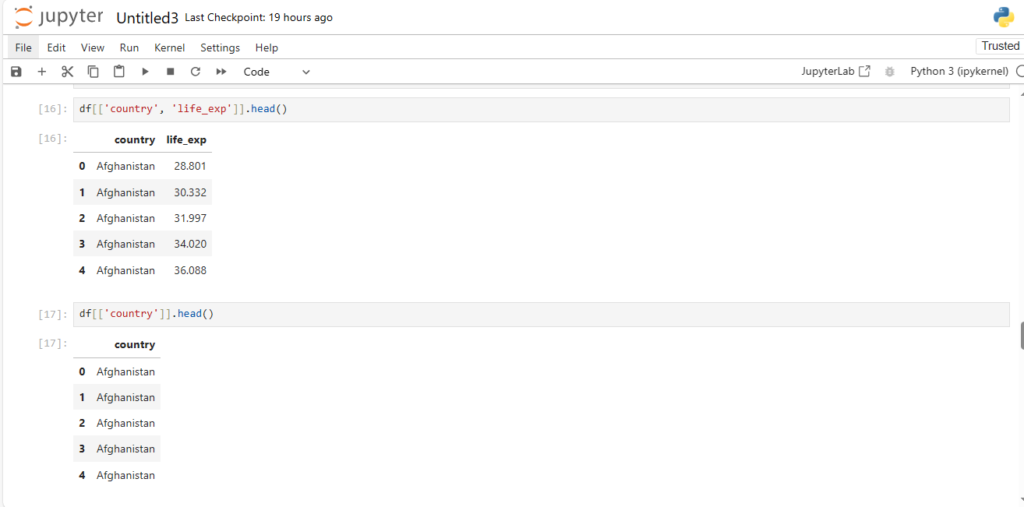
df[['country', 'life_exp']].head()
And what if we pass a single column name?
df[['country']].head()
Note:
Notice how this output type is different from our earlier output using df[‘country’]
==> [‘country’] gives series while [[‘country’]] gives dataframe
Now that we know how to access columns, lets answer some questions
Now what if you also want to check the count of each country in the dataframe?
df['country'].value_counts()
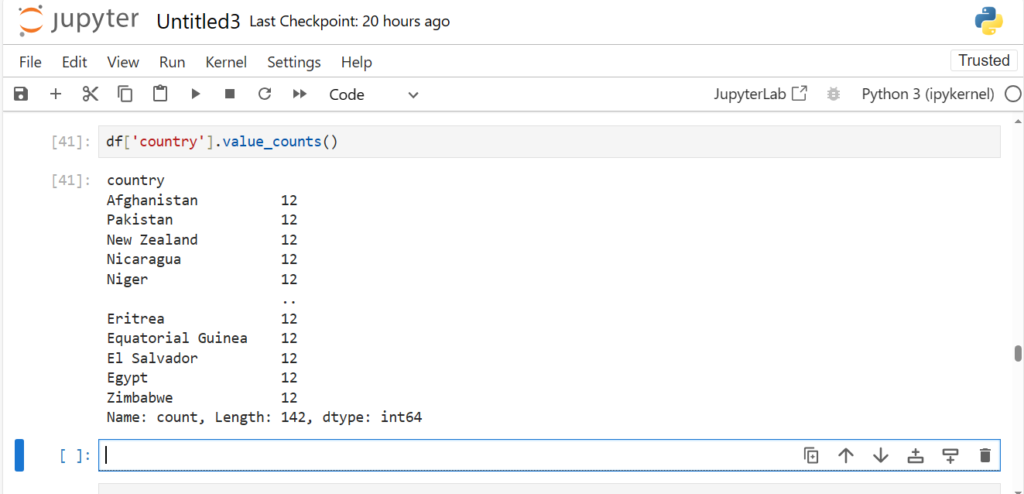
Note:
value_counts() shows the output in decreasing order of frequency
What if we want to change the name of a column ?
We can rename the column by:
- passing the dictionary with old_name:new_name pair
- specifying axis=1
df.rename({"population": "Population", "country":"Country" }, axis = 1) 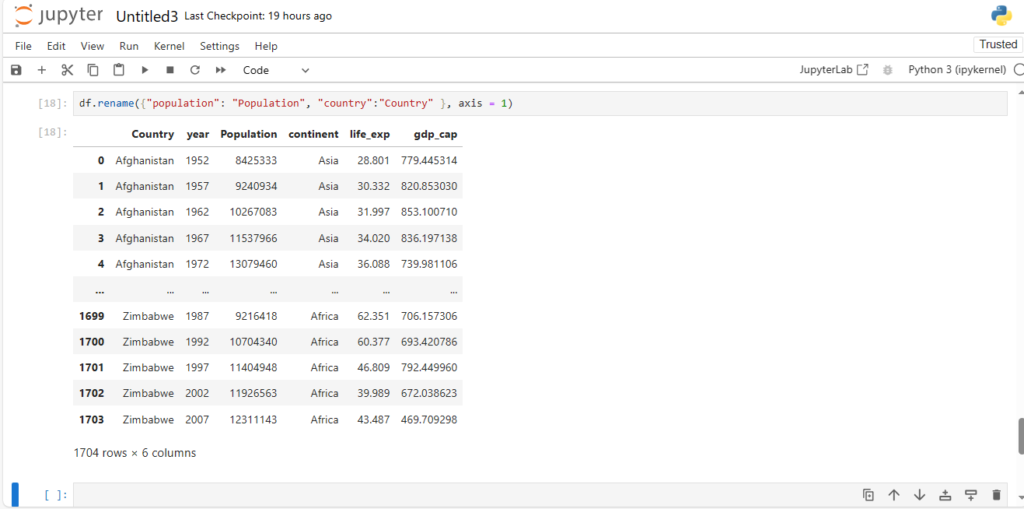
Alternatively, we can also rename the column without using axis
- by using the column parameter
df.rename(columns={"country":"Country"}) 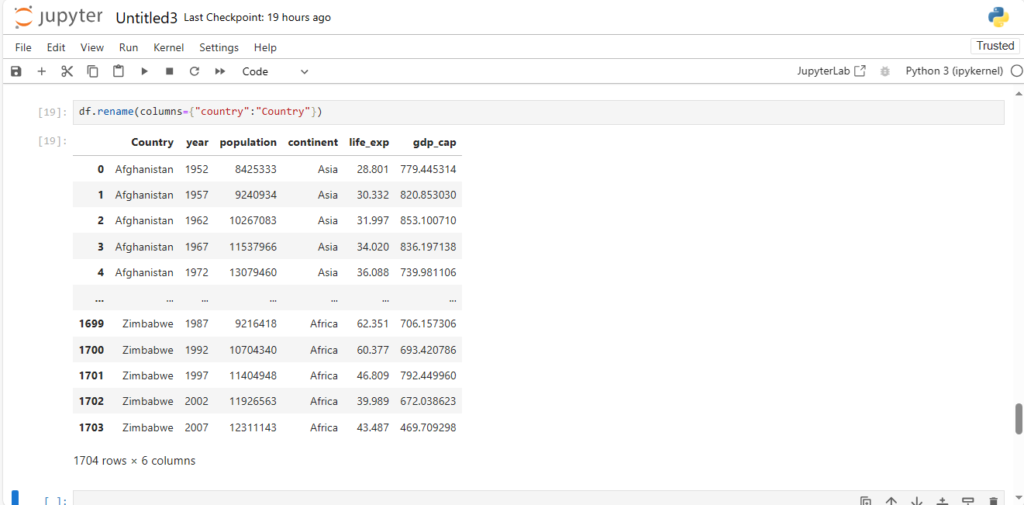
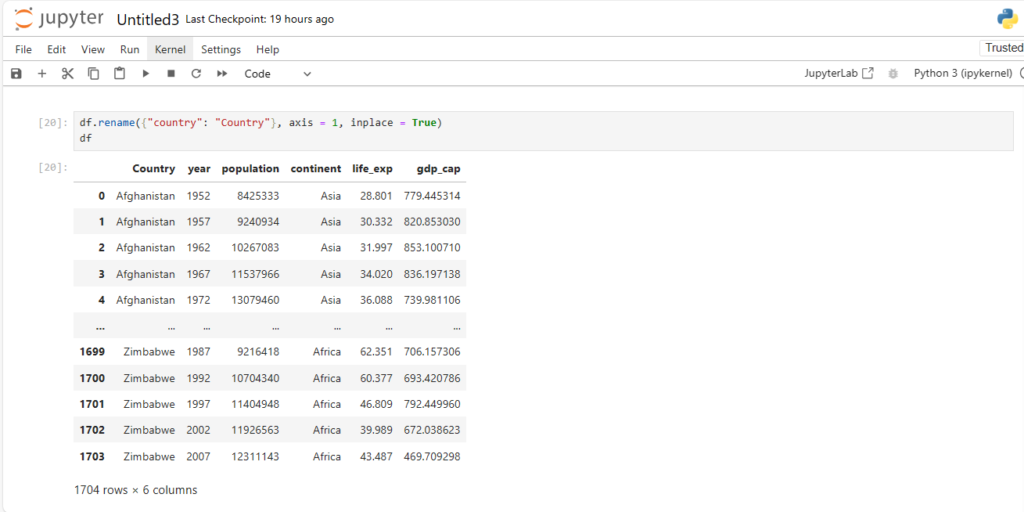
We can set it inplace by setting the inplace argument = True
df.rename({"country": "Country"}, axis = 1, inplace = True)
df Note:
- .rename has default value of axis=0
- If two columns have the same name, then df[‘column’] will display both columns
Now lets try another way of accessing column vals
df.Country
How can we delete cols in pandas dataframe ?
df.drop('continent', axis=1) 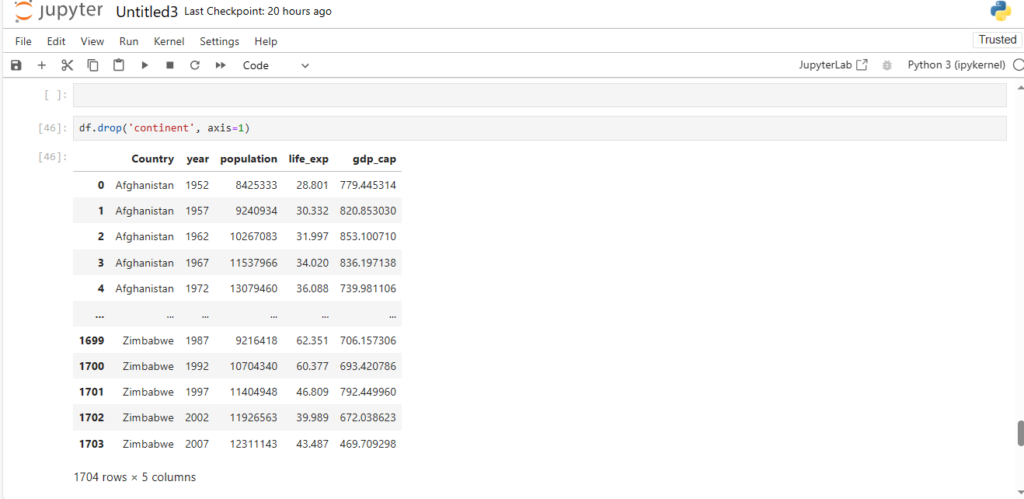
The drop function takes two parameters:
- The column name
- The axis
By default the value of axis is 0
An alternative to the above approach is using the “columns” parameter as we did in rename
df.drop(columns=['continent'])
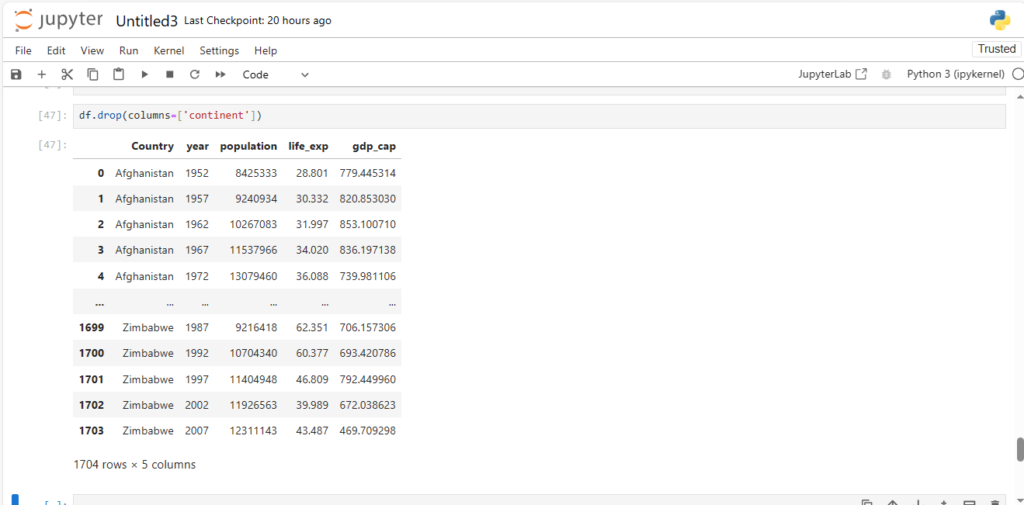
As you can see, column contintent is dropped
Has the column permanently been deleted?
df.head()
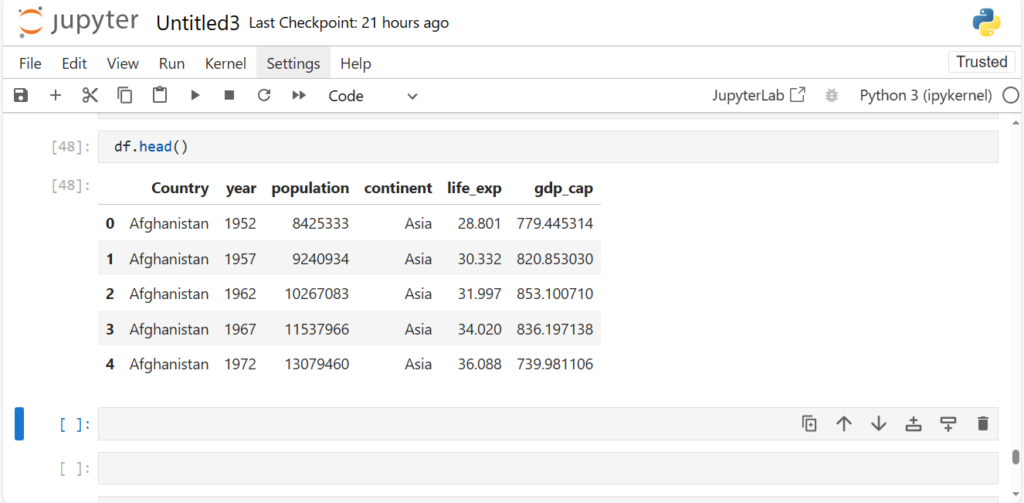
NO, the column continent is still there
Do you see what’s happening here?
We only got a view of dataframe with column continent dropped
How can we permanently drop the column?
We can either re-assign it
- df = df.drop(‘continent’, axis=1)
or
- We can set parameter inplace=True
By default, inplace=False
df.drop('continent', axis=1, inplace=True)df.head() #we print the head to check
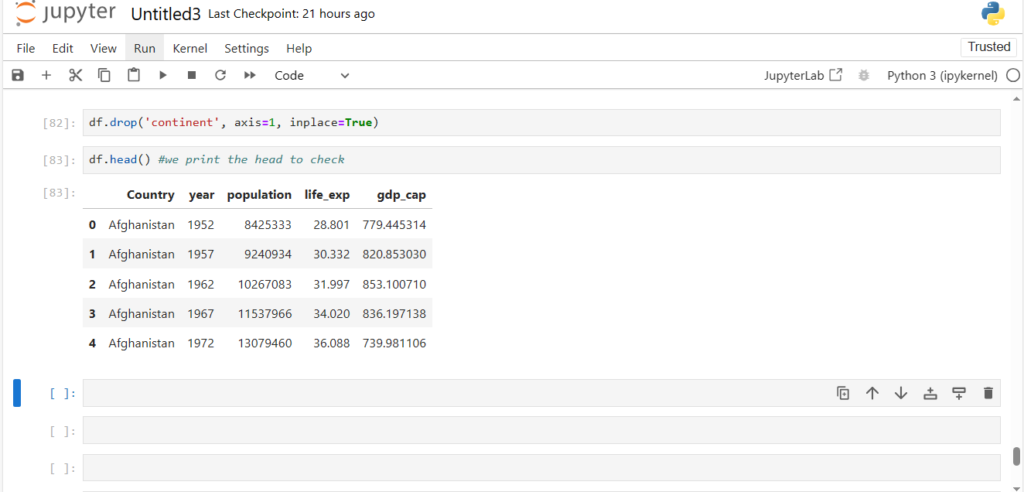
Now we can see the column continent is permanently dropped
Now similarly, what if we want to create a new column?
We can either
- use values from existing columns
or
- create our own values
How to create a column using values from an existing column?
df["year+7"] = df["year"] + 7
df.head()
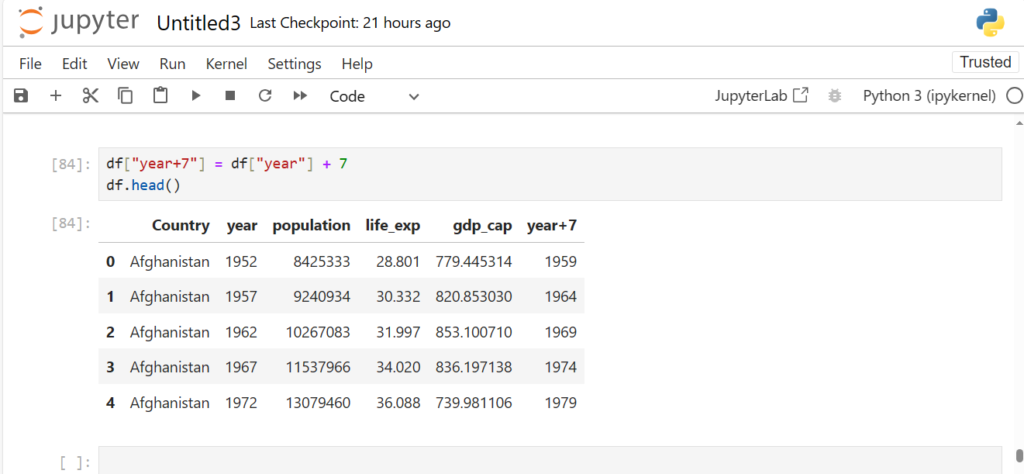
As we see, a new column year+7 is created from the column year
We can also use values from two columns to form a new column
Which two columns can we use to create a new column gdp ?
df['gdp']=df['gdp_cap'] * df['population']
df.head()
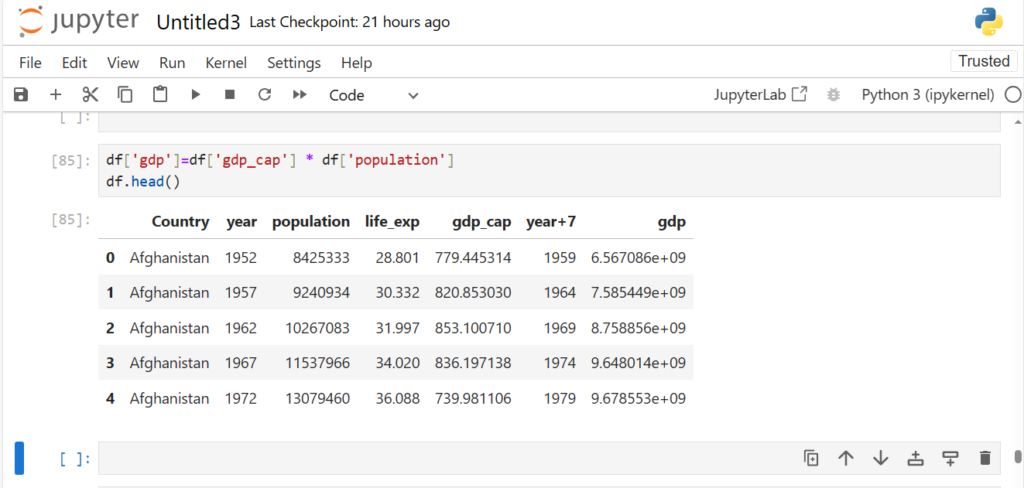
As you can see
- An additional column has been created
- Values in this column are product of respective values in gdp_cap and population
How can we create a new column from our own values?
- We can create a list
OR
- We can create a Pandas Series from a list/numpy array for our new column
df["Own"] = [i for i in range(1704)] # count of these values should be correct
df
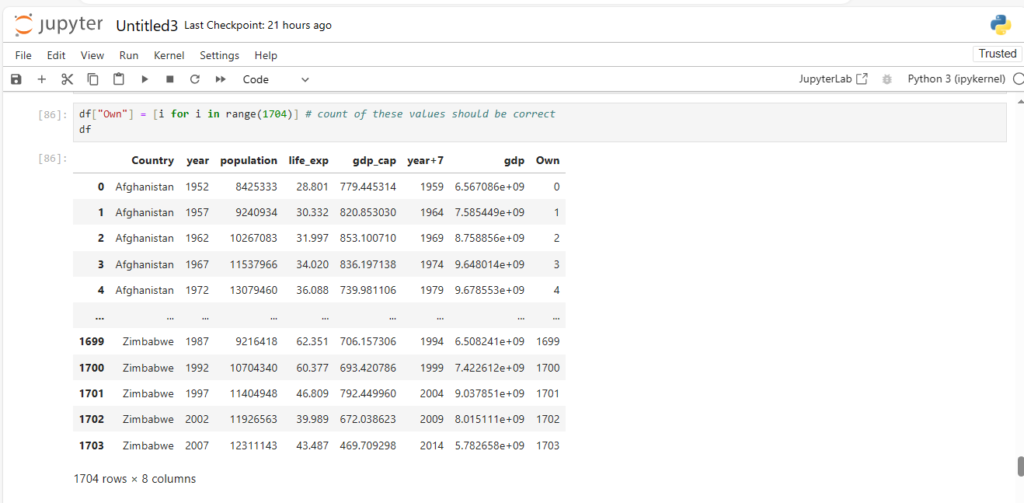
Now that we know how to create new cols lets see some basic ops on rows
Before that lets drop the newly created cols
df.drop(columns=["Own",'gdp', 'year+7'], axis = 1, inplace = True)
df
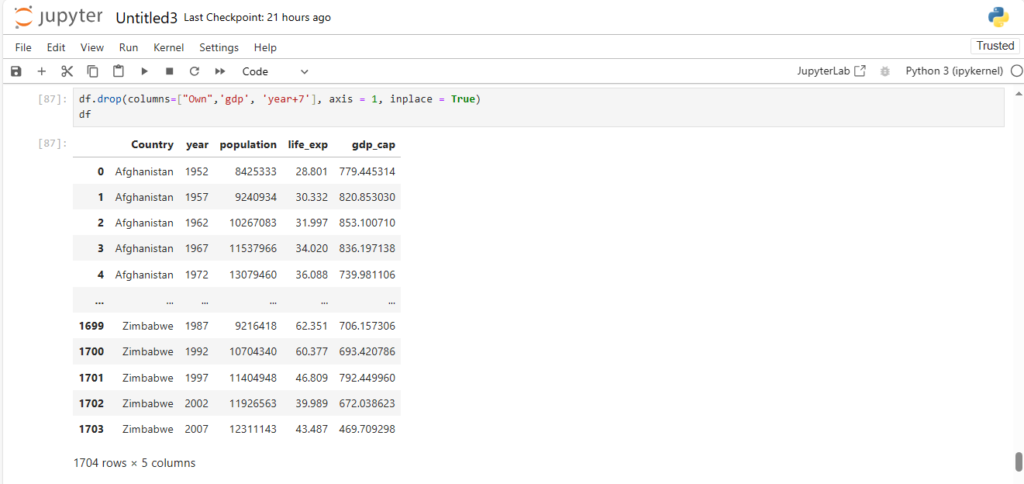
Working with Rows
Notice the indexes in bold against each row
Lets see how can we access these indexes
df.index.values
array([ 0, 1, 2, ..., 1701, 1702, 1703], dtype=int64)
Can we change row labels (like we did for columns)?
What if we want to start indexing from 1 (instead of 0)?
df.index = list(range(1, df.shape[0]+1)) # create a list of indexes of same length
df
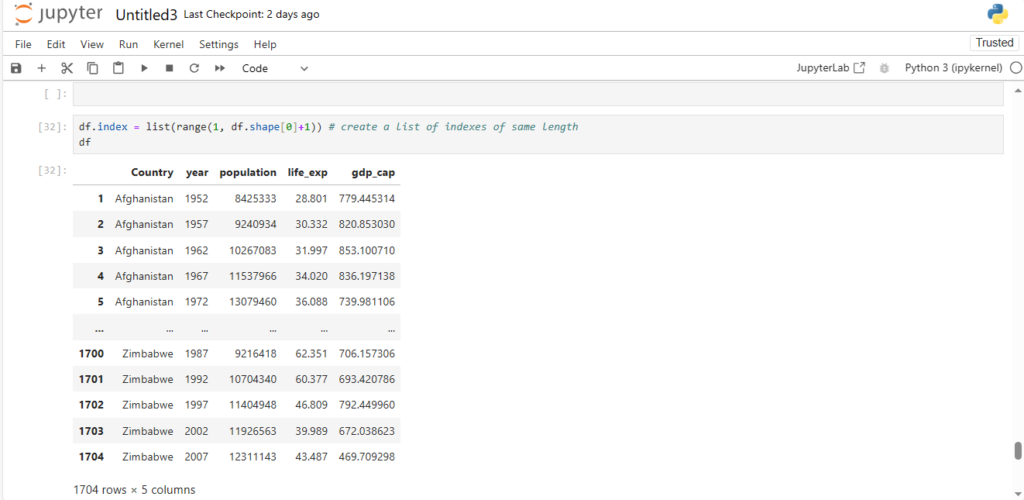
As you can see the indexing is now starting from 1 instead of 0.
Explicit and Implicit Indices
What are these row labels/indices exactly ?
- They can be called identifiers of a particular row
- Specifically known as explicit indices
The python style indices are known as implicit indices
How can we access explicit index of a particular row?
- Using df.index[]
- Takes impicit index of row to give its explicit index
df.index[1] #Implicit index 1 gave explicit index 2
2
But why not use just implicit indexing ?
Explicit indices can be changed to any value of any datatype
- Eg: Explicit Index of 1st row can be changed to First
- Or, something like a floating point value, say 1.0
df.index = np.arange(1, df.shape[0]+1, dtype='float')
df
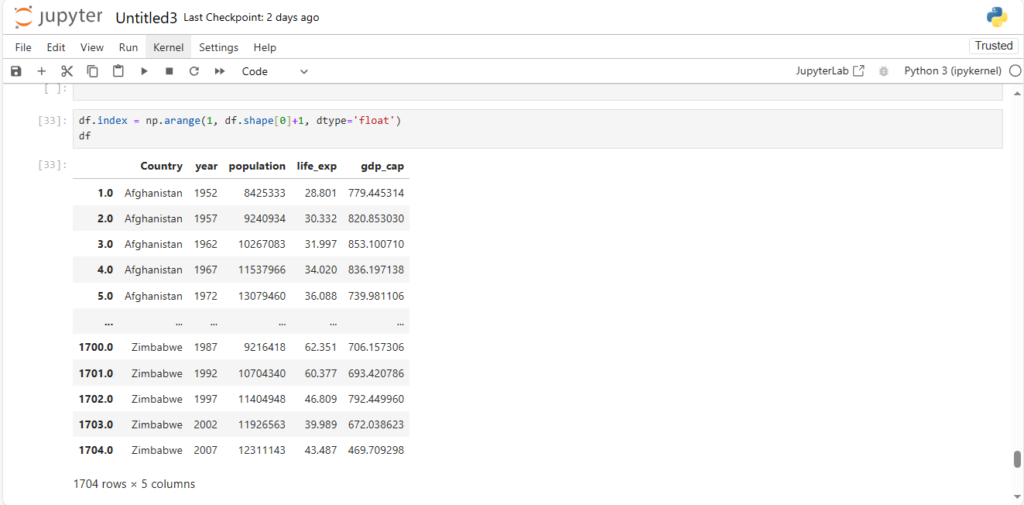
As we can see, the indices are floating point values now
Now to understand string indices, let’s take a small subset of our original dataframe
sample = df.head()
sample
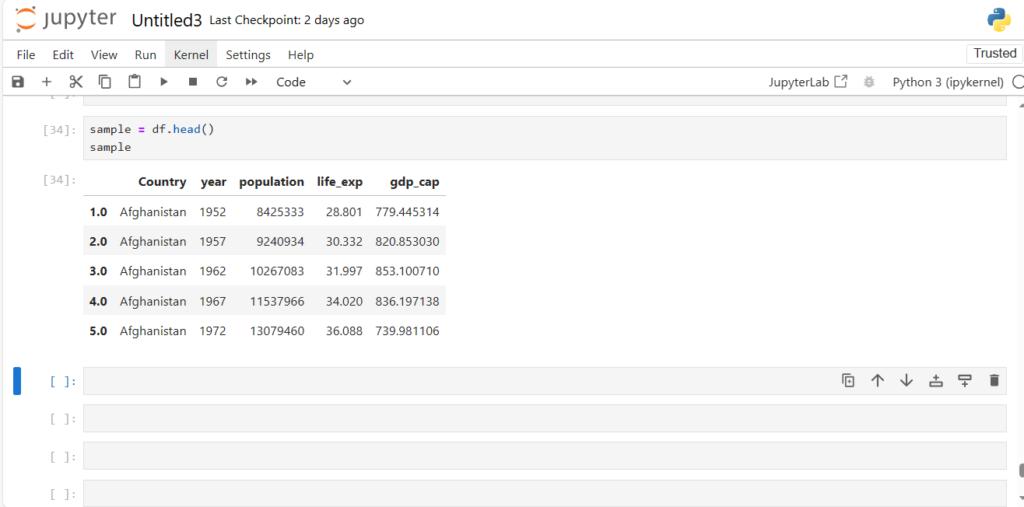
Now what if we want to use string indices?
sample.index = ['a', 'b', 'c', 'd', 'e']
sample
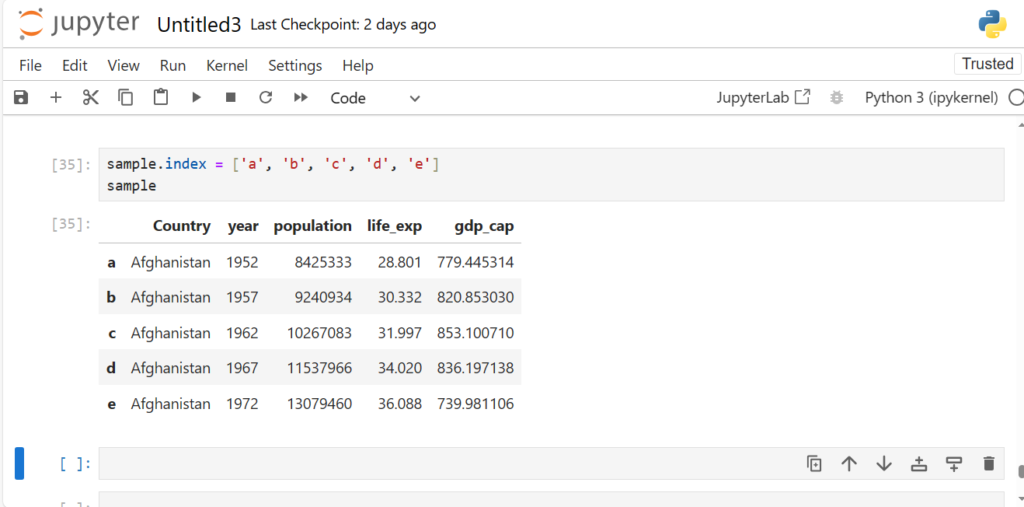
This shows us we can use almost anything as our explicit index
Now let’s reset our indices back to integers
df.index = np.arange(1, df.shape[0]+1, dtype='int')
What if we want to access any particular row (say first row)?
Let’s first see for one column
Later, we can generalise the same for the entire dataframe
ser = df["Country"]
ser.head(20)
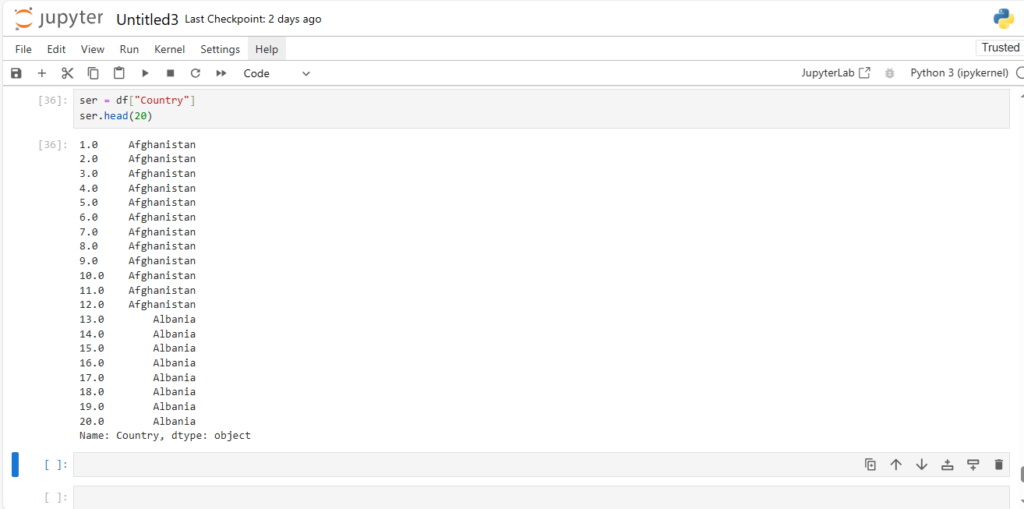
We can simply use its indices much like we do in a numpy array
So, how will be then access the thirteenth element (or say thirteenth row)?
ser[12]
'Afghanistan'
And what about accessing a subset of rows (say 6th:15th) ?
ser[5:15]
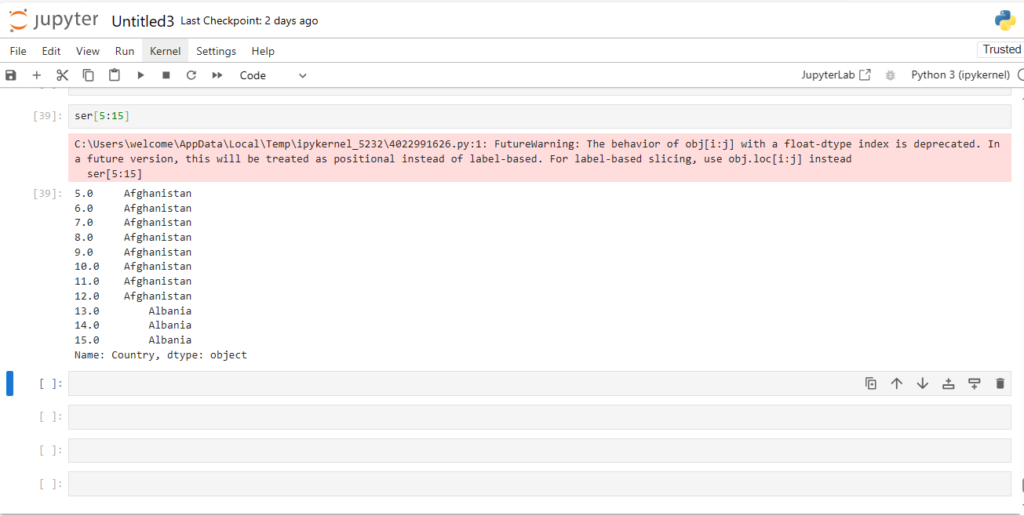
This is known as slicing
Notice something different though?
- Indexing in Series used explicit indices
- Slicing however used implicit indices
How can we access a slice of rows in the dataframe?
df[5:15]
- Woah, so the slicing works
===> Indexing in dataframe looks only for explicit indices \ ===> Slicing, however, checked for implicit
indices - This can be a cause for confusion
- To avoid this pandas provides special indexers, loc and iloc
- We will look at these in a bit Lets look at them one by one
loc and iloc
1. loc
Allows indexing and slicing that always references the explicit index
df.loc[1]
Country Afghanistan
year 1952
population 8425333
life_ex 28.801
gdp_cap 779.445314
Name: 1, dtype: object
df.loc[1:3]
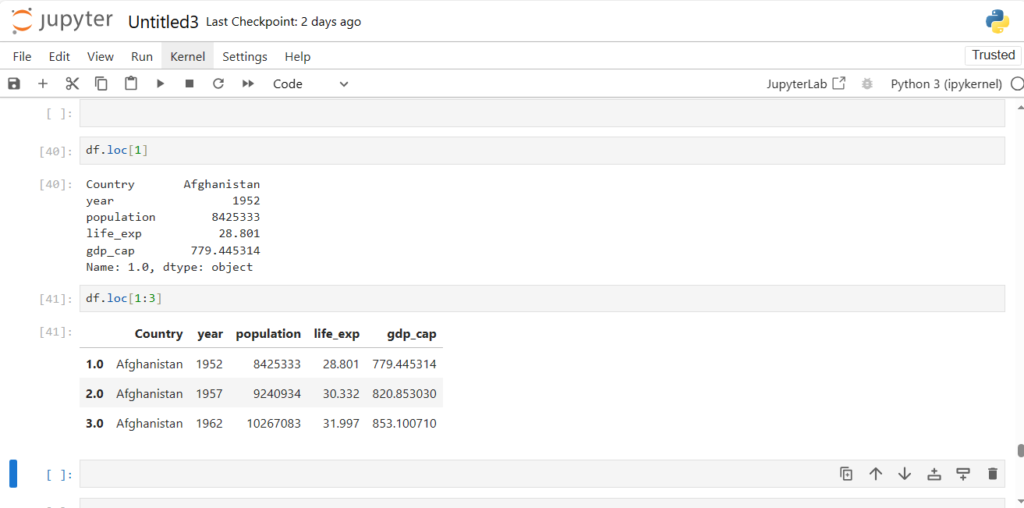
Did you notice something strange here?
- The range is inclusive of end point for loc
- Row with Label 3 is included in the result
2. iloc
Allows indexing and slicing that always references the implicit Python-style index
df.iloc[1]
Country Afghanistan
year 1957
population 9240934
life_exp 30.332
gdp_cap 820.85303
Name: 2, dtype: object
Now will iloc also consider the range inclusive?
df.iloc[0:2]
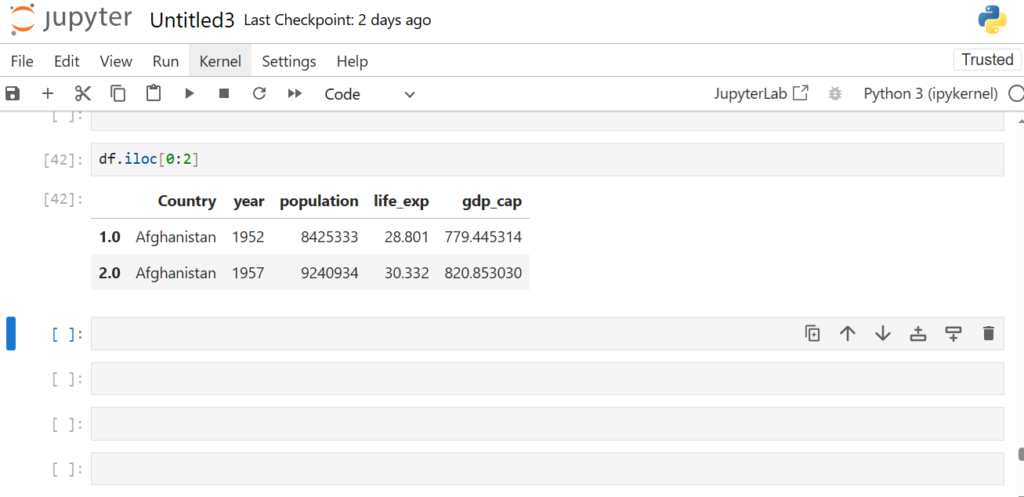
iloc works with implicit Python-style indices
It is important to know about these conceptual differences
Not just b/w loc and iloc , but in general while working in DS and ML
Which one should we use ?
- Generally explicit indexing is considered to be better than implicit
- But it is recommended to always use both loc and iloc to avoid any confusions
What if we want to access multiple non-consecutive rows at same time ?
For eg: rows 1, 10, 100
df.iloc[[1, 10, 100]]
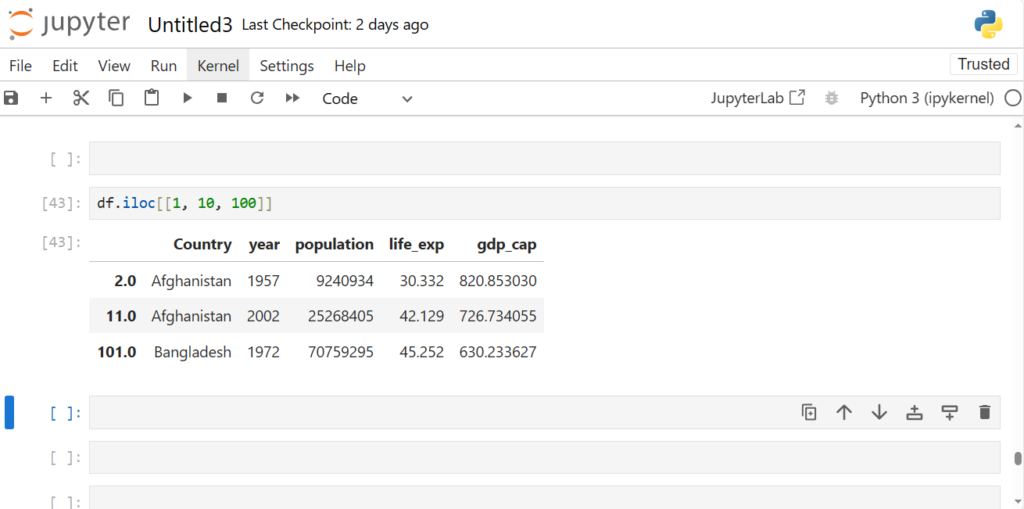
As we see, We can just pack the indices in [] and pass it in loc or iloc
What about negative index?
Which would work between iloc and loc ?
df.iloc[-1]
# Works and gives last row in dataframe
Country Zimbabwe
year 2007
population 12311143
life_exp 43.487
gdp_cap 469.709298
Name: 1704, dtype: object
df.loc[-1]
# Does NOT work
So, why did iloc[-1] worked, but loc[-1] didn’t?
- Because iloc works with positional indices, while loc with assigned labels
- [-1] here points to the row at last position in iloc
Can we use one of the columns as row index?
temp = df.set_index("Country")
temp 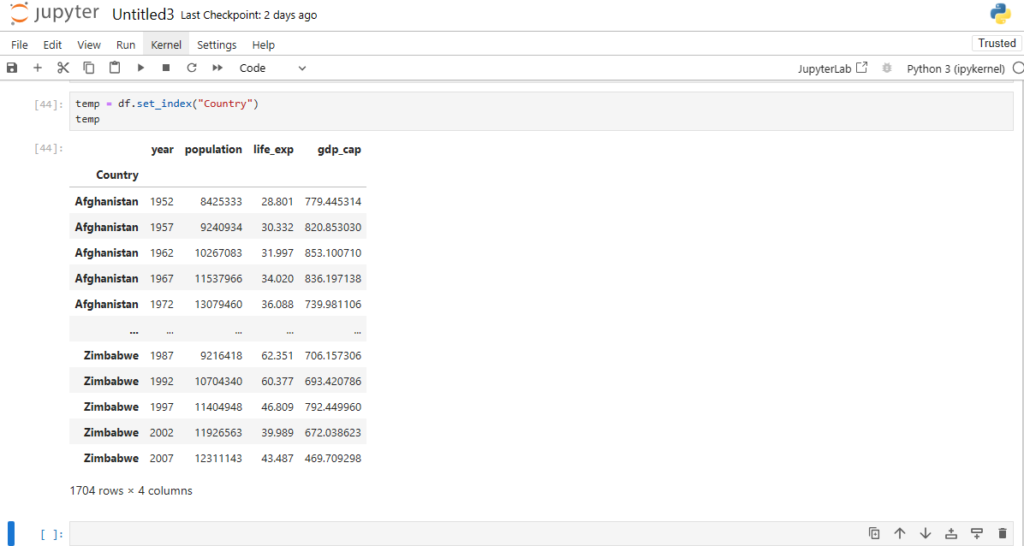
Now what would the row corresponding to index Afghanistan give?
temp.loc['Afghanistan']
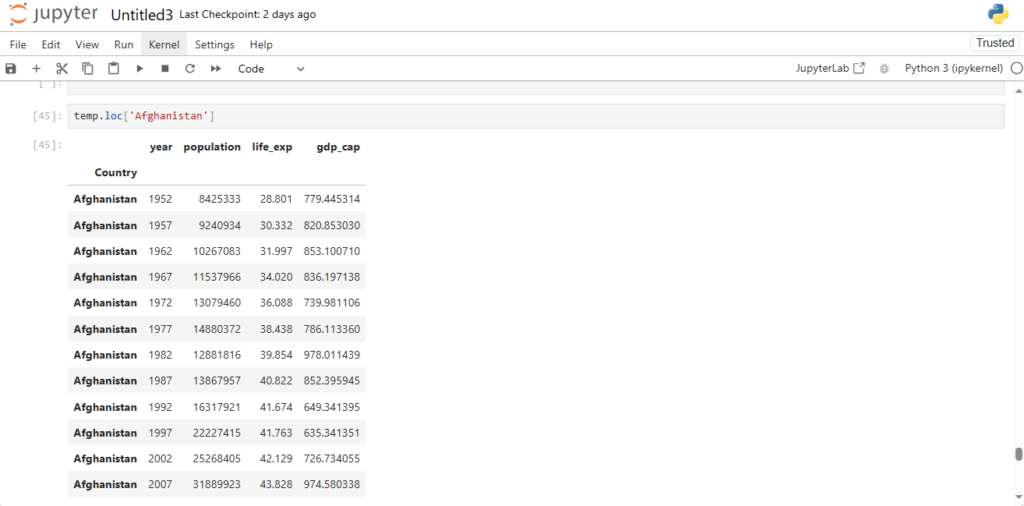
As you can see we got the rows all having index Afghanistan
Now how can we reset our indices back to integers?
df.reset_index()
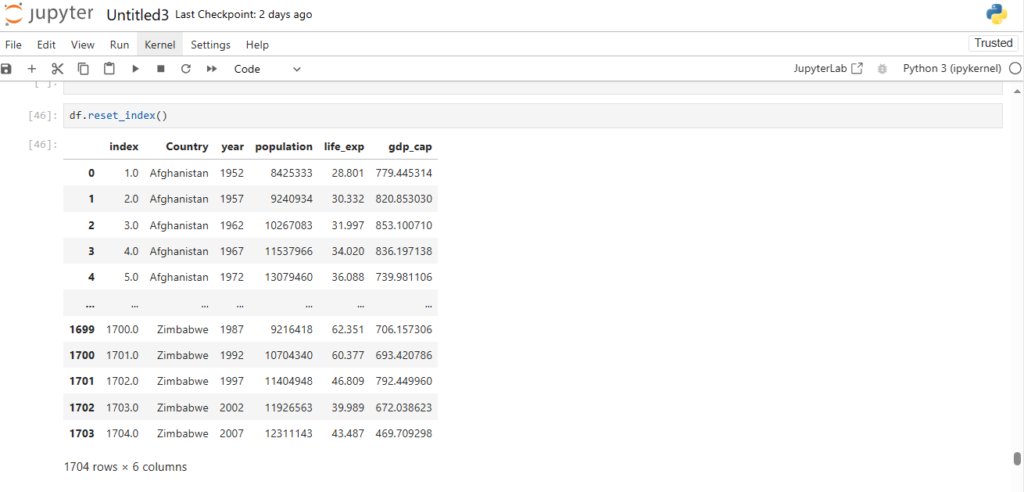
Notice it’s creating a new column index
How can we reset our index without creating this new column?
df.reset_index(drop=True) # By using drop=True we can prevent creation of a new column
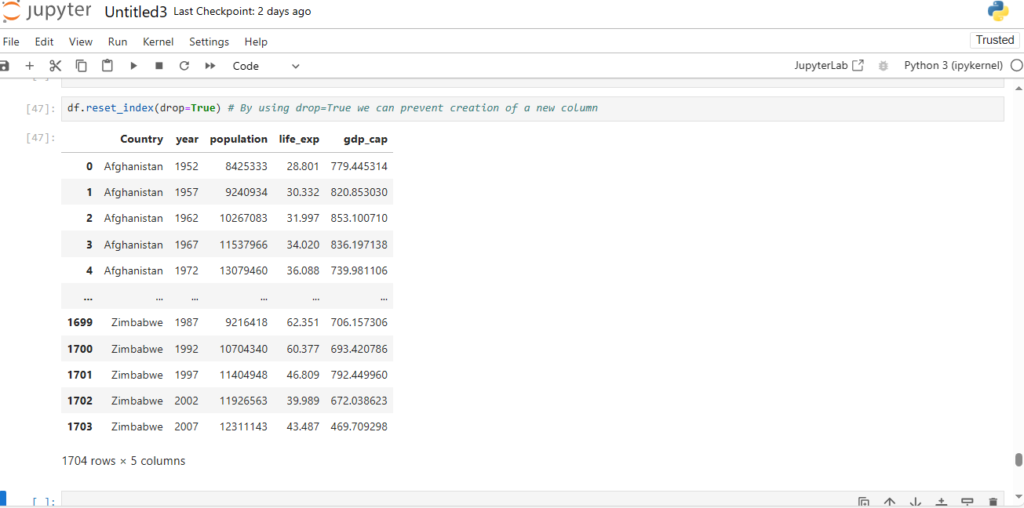
Great, now let’s do this in place
df.reset_index(drop=True, inplace=True)
Now how can we add a row to our dataframe?
There are multiple ways to do this:
- append()
- loc/iloc
How can we do add a row using the append() method?
new_row = {'Country': 'India', 'year': 2000,'life_exp':37.08,'population':13500000,'gdp_
df.append(new_row)Why are we getting an error here?
Its’ saying the ignore_index() parameter needs to be set to True
new_row = {'Country': 'India', 'year': 2000,'life_exp':37.08,'population':13500000,'gdp_
df = df.append(new_row, ignore_index=True)
df
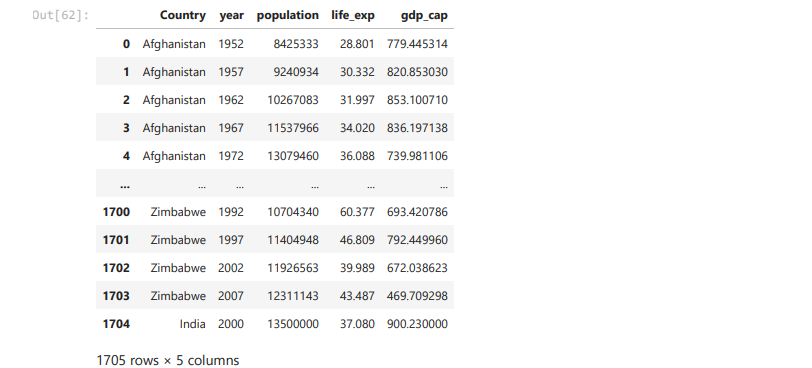
What you can infer from last two duplicate rows ?
Dataframe allow us to feed duplicate rows in the data
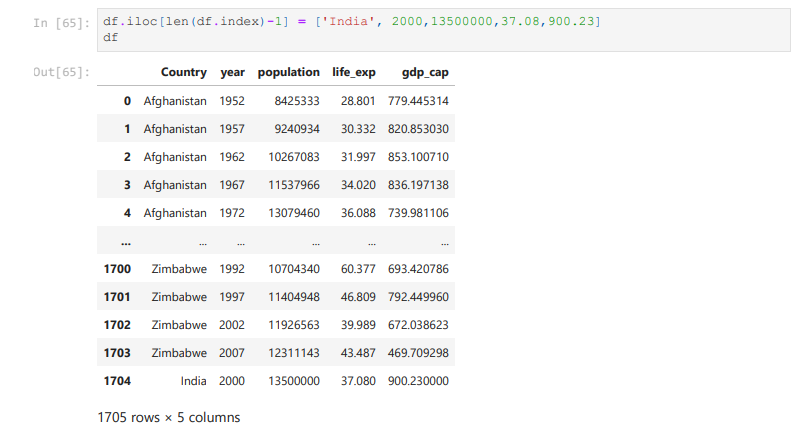
Now, can we also use iloc?
Adding a row at a specific index position will replace the existing row at that position.
df.iloc[len(df.index)-1] = ['India', 2000,13500000,37.08,900.23]
df
Now what if we want to delete a row ?
Use df.drop()
If you remember we specified axis=1 for columns
We can modify this for rows
- We can use axis=0 for rows
Does drop() method uses positional indices or labels?
What do you think by looking at code for deleting column?
- We had to specify column title
- So drop() uses labels, NOT positional indices
# Let's drop row with label 3
df = df.drop(3, axis=0)
df
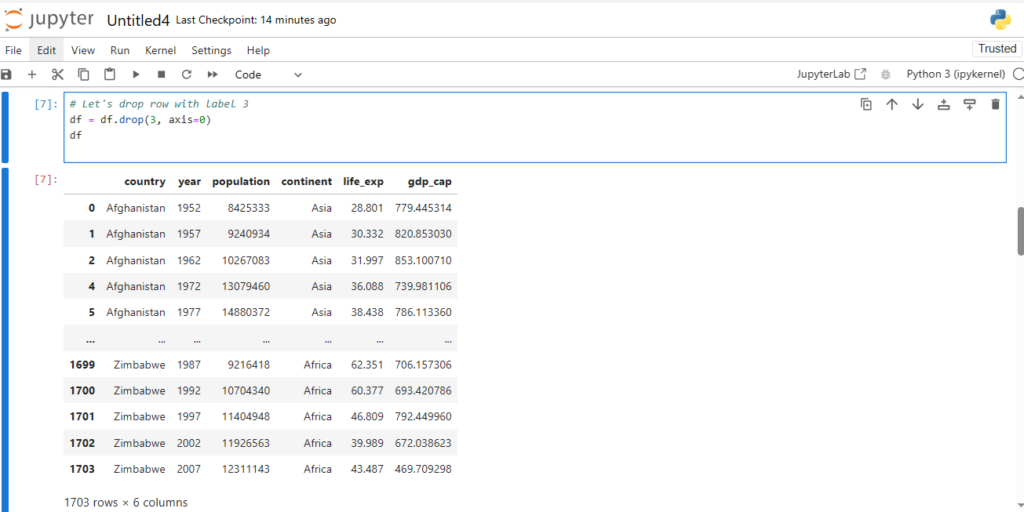
Now we see that row with label 3 is deleted
We now have rows with labels 0, 1, 2, 4, 5, …
Now df.loc[4] and df.iloc[4] will give different rows
df.loc[4] # The 4th row is printed
Country Afghanistan
year 1972
population 13079460
life_exp 36.088
gdp_cap 739.981106
Name: 4, dtype: object
df.iloc[4] # The 5th row is printed
Country Afghanistan
year 1977
population 14880372
life_exp 38.438
gdp_cap 786.11336
Name: 5, dtype: object
And hww can we drop multiple rows?
df.drop([1, 2, 4], axis=0) # drops rows with labels 1, 2, 4
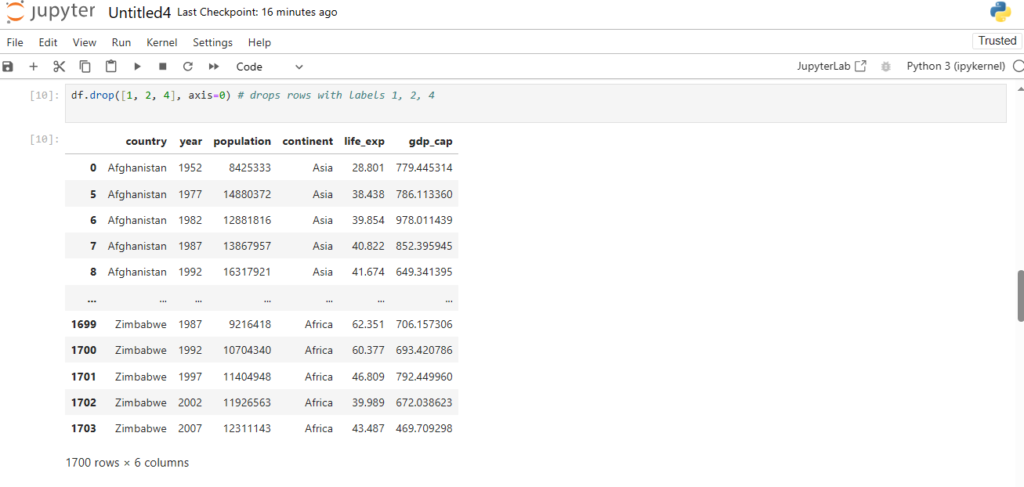
Let’s reset our indices now
df.reset_index(drop=True,inplace=True)
Now if you remember, the last two rows were duplicates.
How can we deal with these duplicate rows?
Let’s create some more duplicate rows to understand this
df.loc[len(df.index)] = ['India',2000,13500000,37.08,900.23]
df.loc[len(df.index)] = ['Sri Lanka',2022 ,130000000,80.00,500.00]
df.loc[len(df.index)] = ['Sri Lanka',2022 ,130000000,80.00,500.00]
df.loc[len(df.index)] = ['India',2000 ,13500000,80.00,900.23]
df
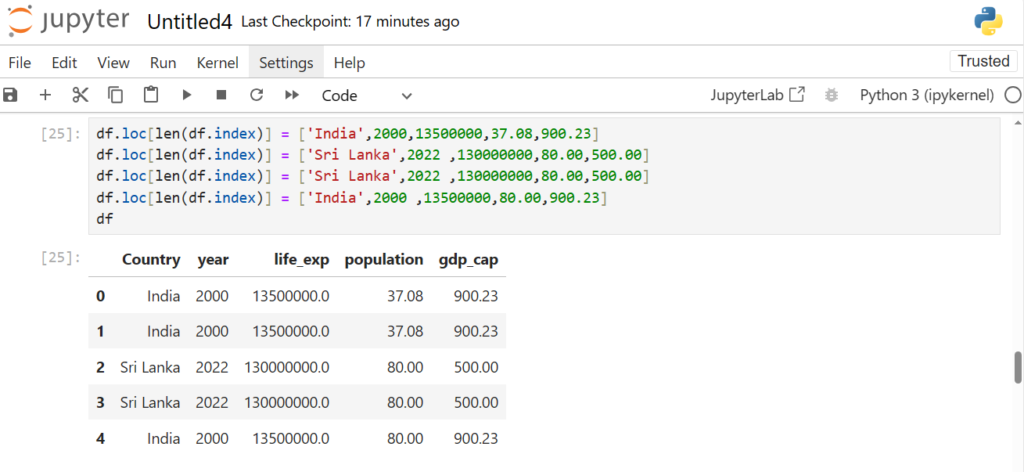
Now how can we check for duplicate rows?
Use duplicated() method on the DataFrame
df.duplicated()
0 False
1 False
2 False
3 False
4 False
...
1703 False
1704 True
1705 False
1706 True
1707 False
Length: 1708, dtype: bool
It outputs True if an entire row is identical to a previous row.
However, it is not practical to see a list of True and False
We can Pandas loc data selector to extract those duplicate rows
# Extract duplicate rows
df.loc[df.duplicated()]
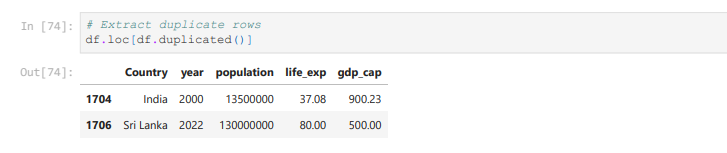
Working with Rows and Columns together
How can we slice the dataframe into, say, first 4 rows and first 3 columns?
We can use iloc
df.iloc[1:5, 1:4]
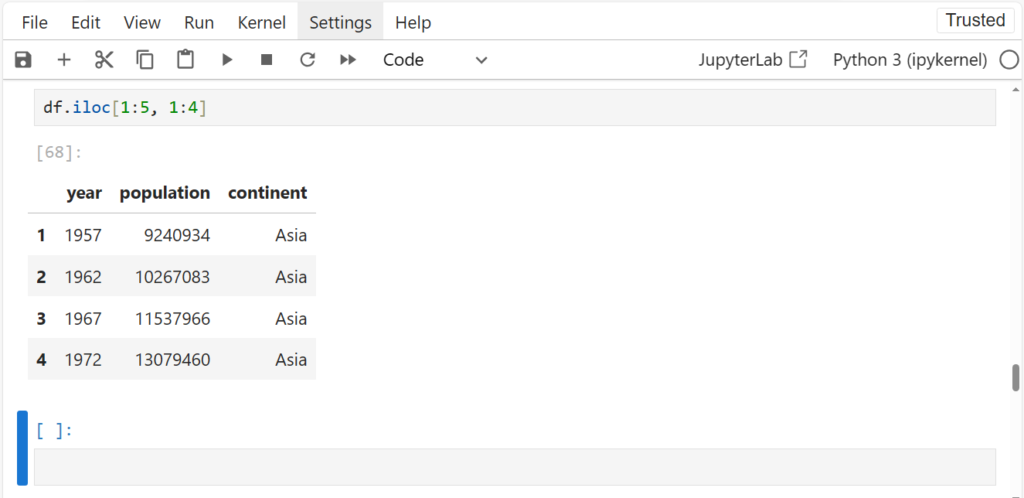
Pass in 2 different ranges for slicing – one for row and one for column just like Numpy
Recall, iloc doesn’t include the end index while slicing
We can mention ranges using column labels as well in loc
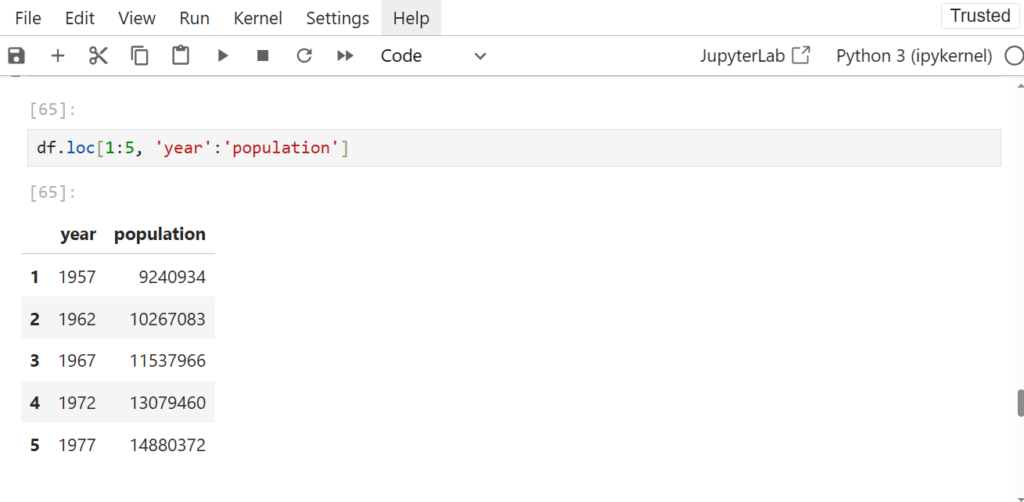
df.loc[1:5, 'year':'population']
How can we get specific rows and columns?
df.iloc[[0,10,100], [0,2,3]]
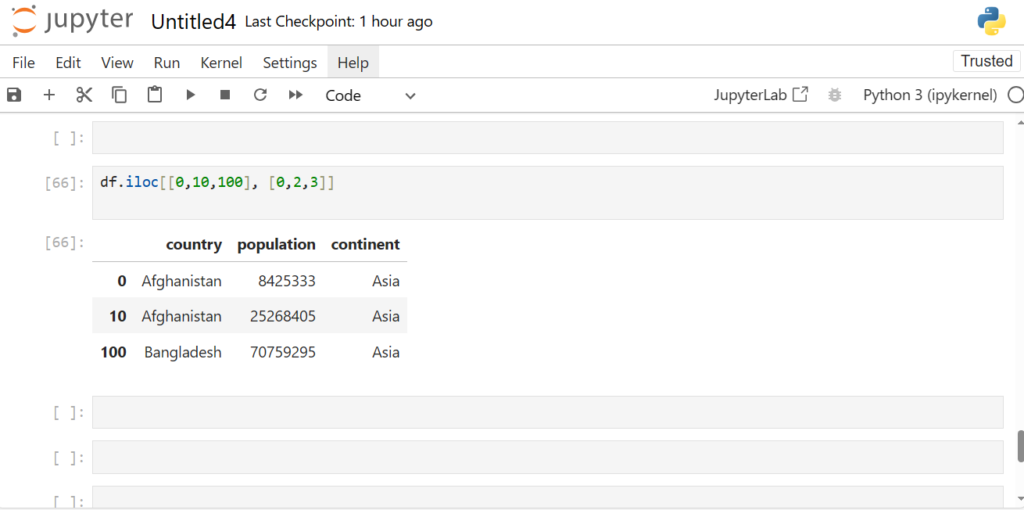
We pass in those specific indices packed in []
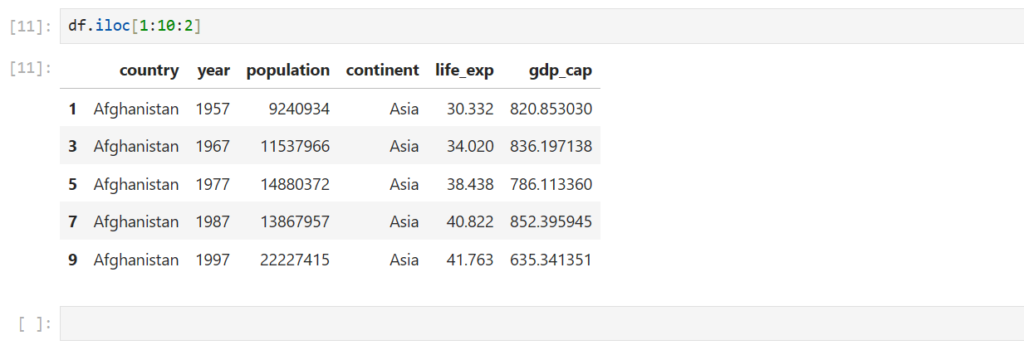
Can we do step slicing?
Yes, just like we did in Numpy
df.iloc[1:10:2]
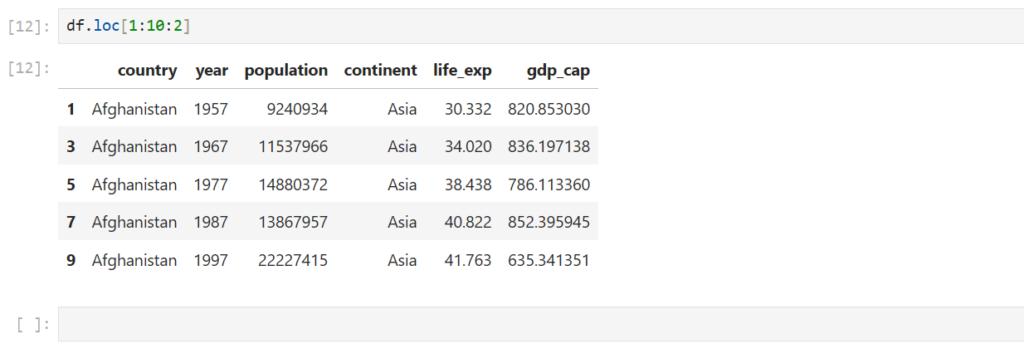
Does step slicing work for loc too?
df.loc[1:10:2]
Pandas built-in operation
Let’s select the feature ‘life_exp‘
le = df['life_exp']
le
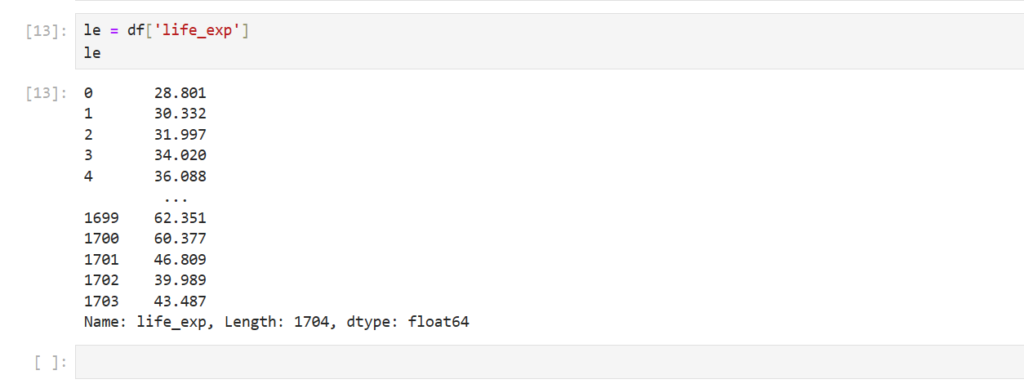
How can we find the mean of the col life_exp ?
le.mean()
=> 59.499171358313774
What other operations can we do?
- sum()
- count()
- min()
- max()
… and so on
Note:
We can see more methods by pressing “tab” after le.
le.sum()
=> 101624.58468
le.count()
=>1708
What will happen we get if we divide sum() by count() ?
le.sum() / le.count()
=> 59.499171358313816
It gives the mean of life expectancy
Sorting
If you notice, life_exp col is not sorted
How can we perform sorting in pandas ?
df.sort_values(['life_exp'])
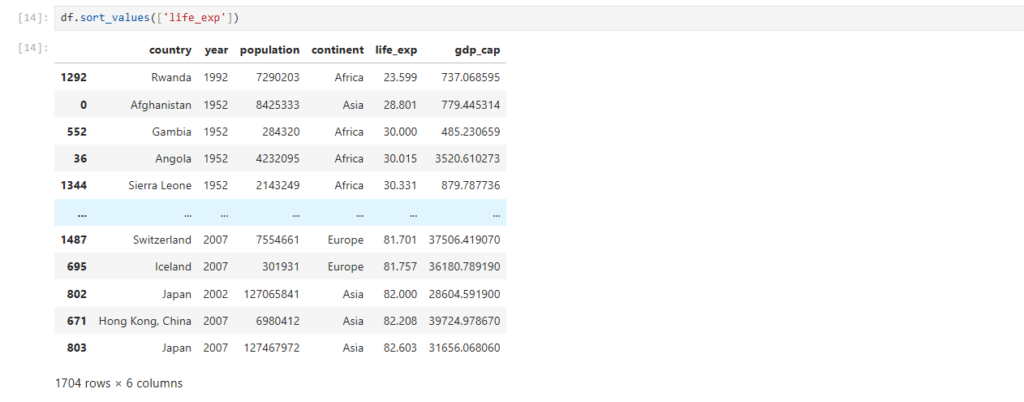
Rows get sorted based on values in life_exp column
By default, values are sorted in ascending order
How can we sort the rows in descending order?
df.sort_values(['life_exp'], ascending=False)
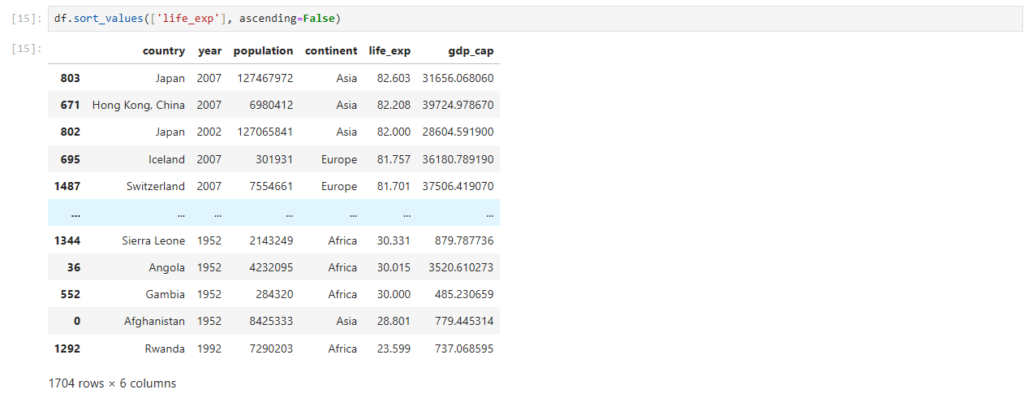
Now the rows are sorted in descending
Can we do sorting on multiple columns?
df.sort_values(['year', 'life_exp'])
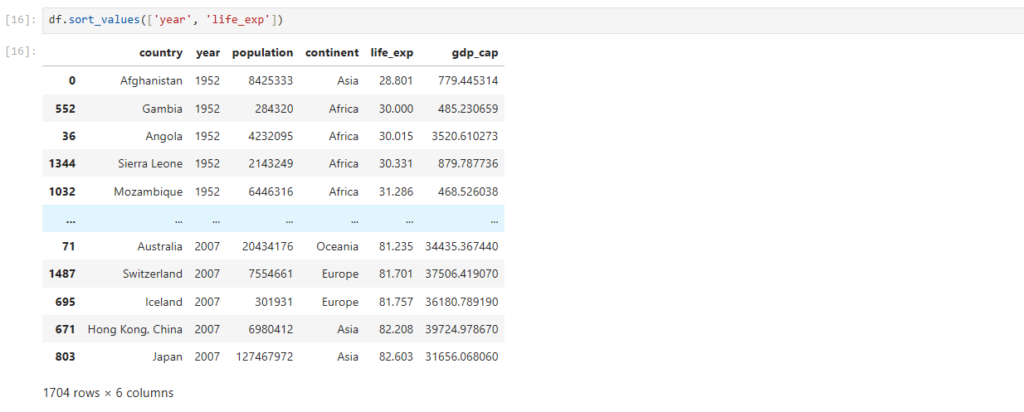
What exactly happened here?
- Rows were first sorted based on ‘year‘
- Then, rows with same values of ‘year’ were sorted based on ‘lifeExp‘
This way, we can do multi-level sorting of our data?
How can we have different sorting orders for different columns in multi-level sorting?
df.sort_values(['year', 'life_exp'], ascending=[False, True])
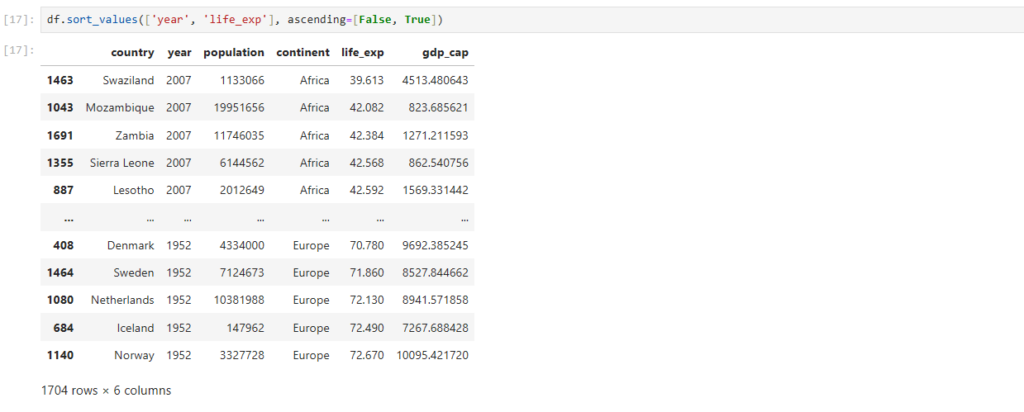
Just pack True and False for respective columns in a list []
Concatenating DataFrames
Let’s use a mini use-case of users and messages
users –> Stores the user details – IDs and Names of users
users = pd.DataFrame({"userid":[1, 2, 3], "name":["kiran", "alok", "vikas"]})
users 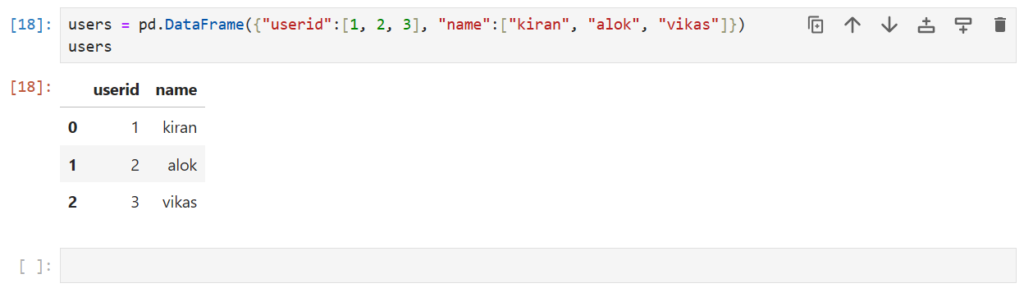
msgs –> Stores the messages users have sent – User IDs and messages
msgs = pd.DataFrame({"userid":[1, 1, 2, 4], "msg":['ok', "thik", "wow", "hmm"]})
msgs 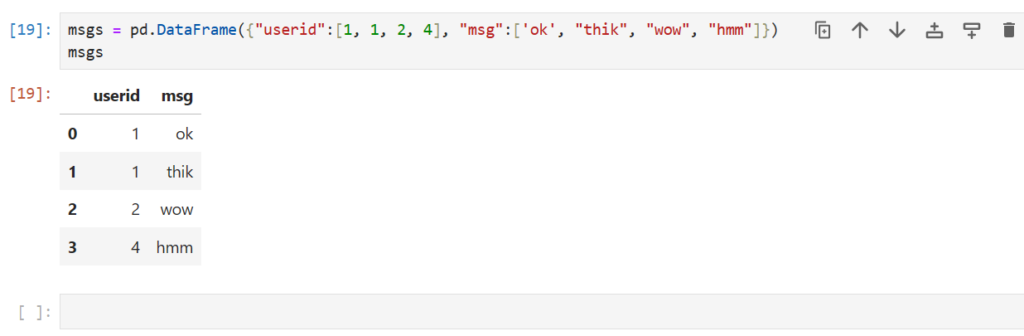
Can we combine these 2 DataFrames to form a single DataFrame?
pd.concat([users, msgs])
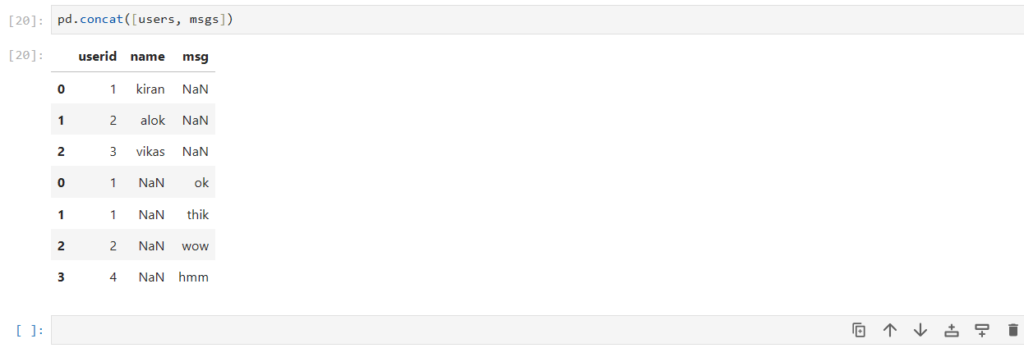
How exactly did concat work?
- By default, axis=0 (row-wise) for concatenation
- userid , being same in both DataFrames, was combined into a single column
- First values of users dataframe were placed, with values of column msg as NaN
- Then values of msgs dataframe were placed, with values of column msg as NaN
- The original indices of the rows were preserved
Now how can we make the indices unique for each row?
pd.concat([users, msgs], ignore_index = True)
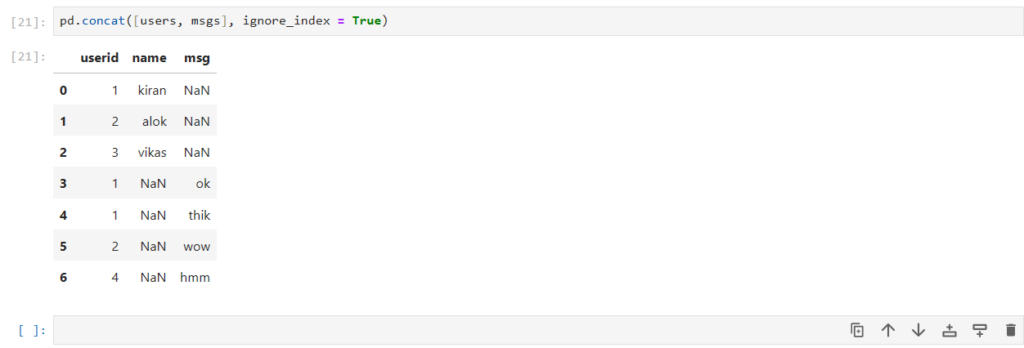
How can we concatenate them horizontally?
pd.concat([users, msgs], axis=1)
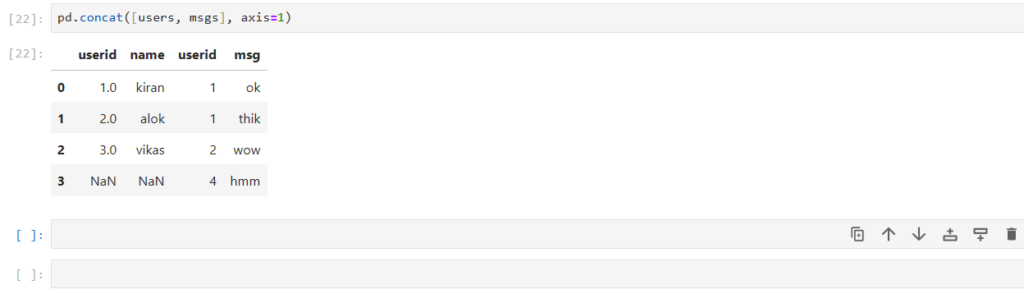
As you can see here:
- Both the dataframes are combined horizontally (column-wise)
- It gives 2 columns with different positional (implicit) index, but same label
Merging Dataframes
So far we have only concatenated and not merged data
But what is the difference between concat and merge ?
- concat
simply stacks multiple DataFrame together along an axis
- merge
combines dataframes in a smart way based on values in shared columns
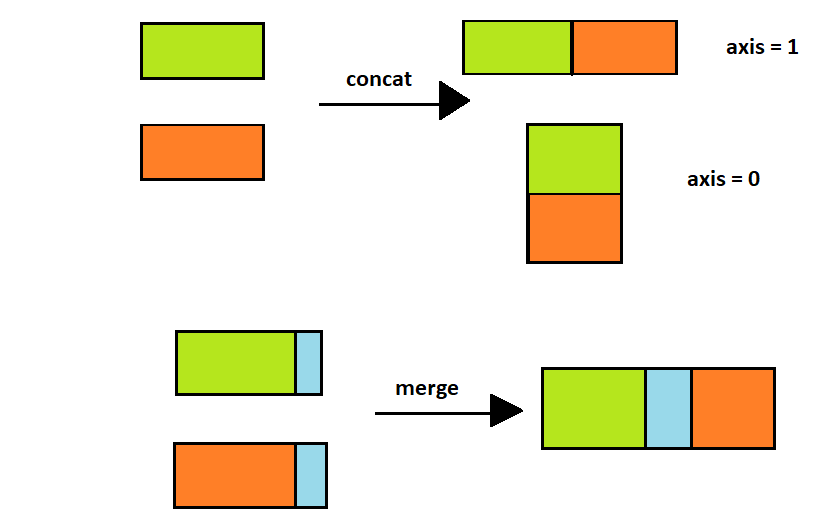
How can we know the name of the person who sent a particular message?
We need information from both the dataframes
So can we use pd.concat() for combining the dataframes ?
NO
pd.concat([users, msgs], axis=1)
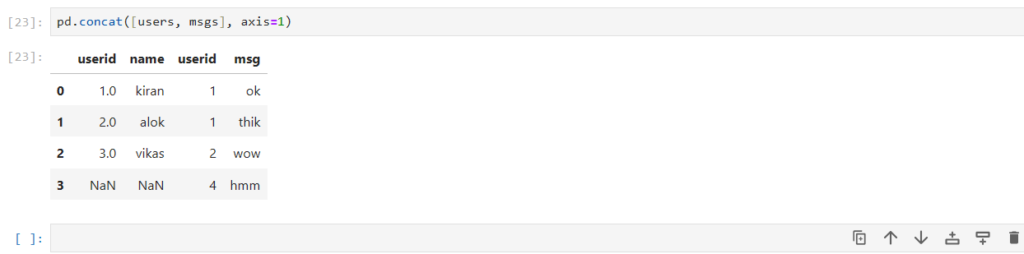
What are the problems with concat here?
- concat simply combined/stacked the dataframe horizontally
- If you notice, userid 3 for user dataframe is stacked against userid 2 for msg dataframe
- This way of stacking doesn’t help us gain any insights
=> pd.concat() does not work according to the values in the columns
We need to merge the data
How can we join the dataframes ?
users.merge(msgs, on="userid")
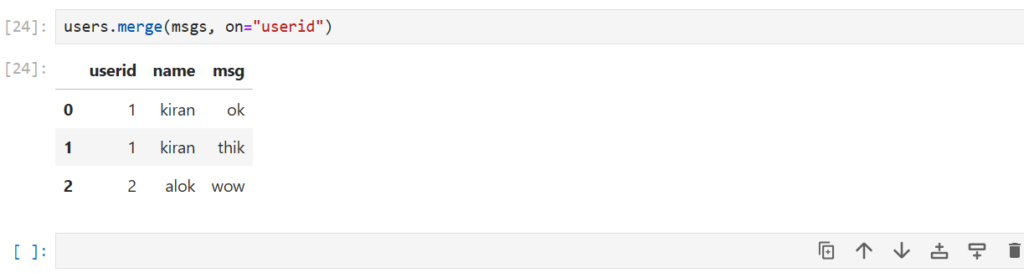
Notice that users has a userid = 3 but msgs does not
- When we merge these dataframes the userid = 3 is not included
- Similarly, userid = 4 is not present in users , and thus not included
- Only the userid common in both dataframes is shown
What type of join is this?
Inner Join
Remember joins from SQL?
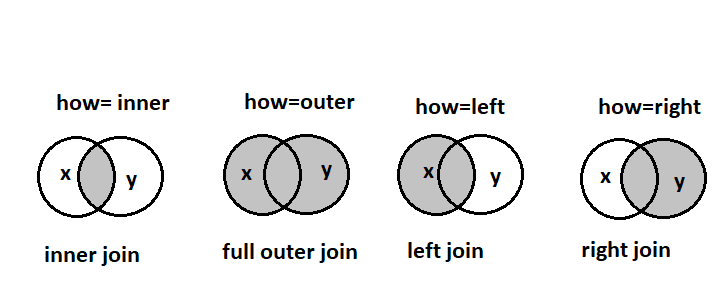
The on parameter specifies the key , similar to primary key in SQL
Now what join we want to use to get info of all the users and all the messages?
users.merge(msgs, on = "userid", how="outer")
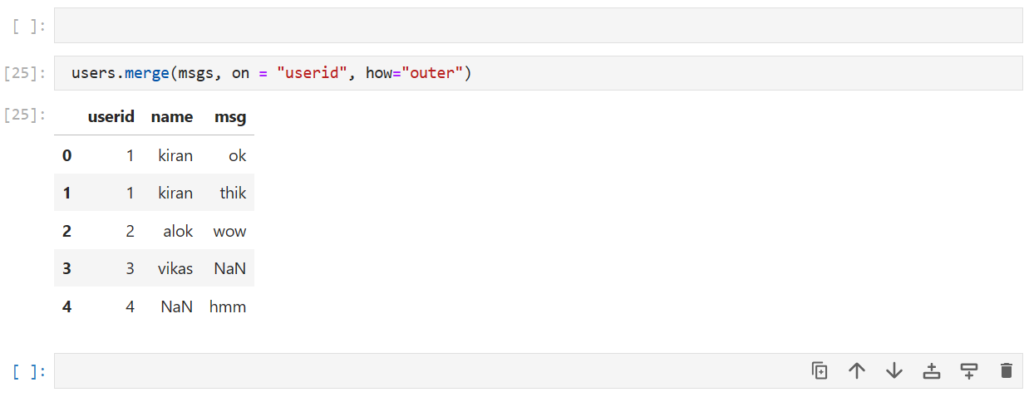
Note:
All missing values are replaced with NaN
And what if we want the info of all the users in the dataframe?
users.merge(msgs, on = "userid",how="left")
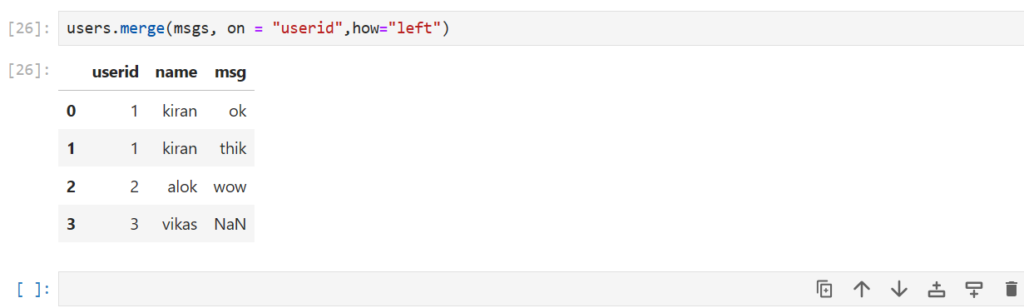
what if we want all the messages and info only for the users who sent a message?
users.merge(msgs, on = "userid", how="right")
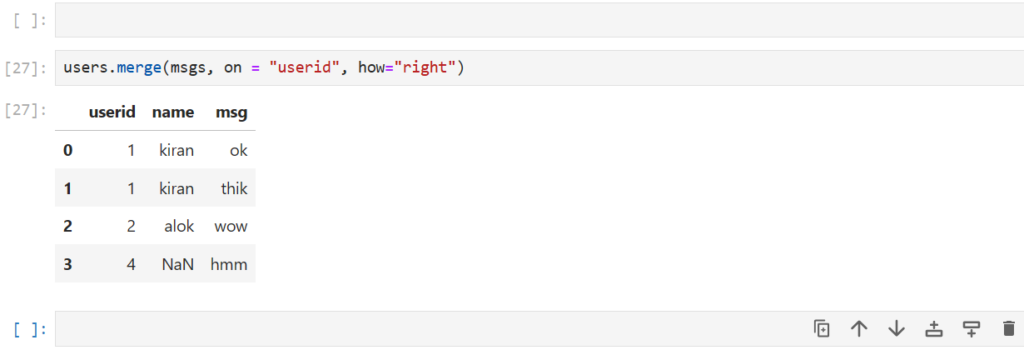
Note,
NaN in name can be thought of as an anonymous message
But sometimes the column names might be different even if they contain the same data
Let’s rename our users column userid to id
users.rename(columns = {"userid": "id"}, inplace = True)
users 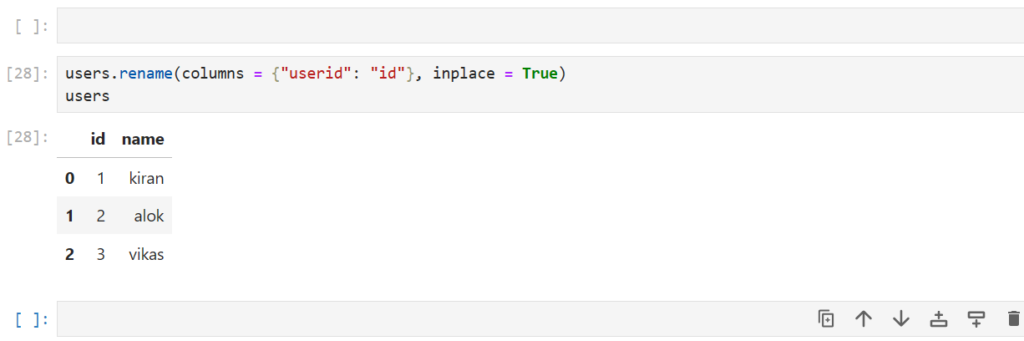
how can we merge the 2 dataframes when the key has a different name ?
users.merge(msgs, left_on="id", right_on="userid")
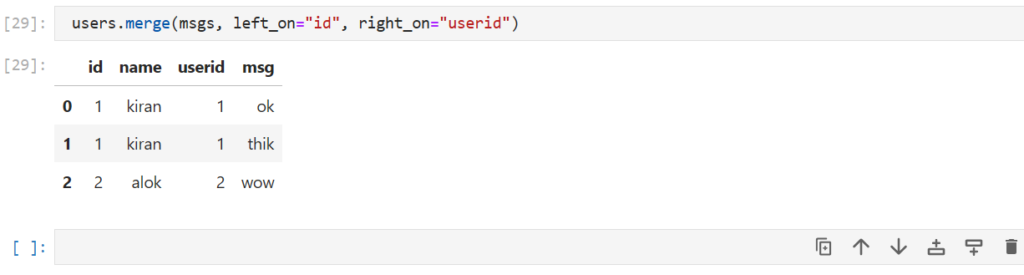
Here,
- left_on : Specifies the key of the 1st dataframe (users here)
- right_on : Specifies the key of the 2nd dataframe (msgs here)
