If you are starting your journey in data cleaning with Python, the first step is understanding what Pandas in Python really is. In this section, we explain the basics of Pandas, DataFrame (df), and why it is the most popular tool for data manipulation and data cleansing in Python.
1. What is Pandas in Python?
Answer: Pandas is an open-source Python library used for
data analysis, cleaning, and manipulation. It provides flexible data
structures like Series (1-D) and DataFrame (2-D) to work with
structured data (tables, CSVs, Excel, SQL queries).
2. What is Pandas Python used for?
Answer: Pandas is mainly used for:
- ✓ Data Cleaning (handling missing values, duplicates)
- ✓ Data Transformation (filtering, grouping, sorting)
- ✓ Data Analysis (statistics, summarization)
- ✓ Reading/Writing files like CSV, Excel, SQL
3. What are Pandas in Python?
Answer: In simple terms, Pandas are powerful Python tools that help turn raw data into clean, structured tables so analysts can focus on insights instead of messy datasets.
4. What is df in Python?
Answer: In Pandas, df is a common variable name used for
a DataFrame. A DataFrame is a 2D table of rows and columns, just like an
Excel sheet or SQL table.
import pandas as pd
df = pd.read_csv("data.csv")
print(df.head()) # displays first 5 rows
5. What is the difference between Pandas and df?
Answer: Pandas is the library, while df
(DataFrame) is the data structure created using Pandas to store and
manipulate tabular data.
6. Why use Pandas for data cleaning?
Answer: Pandas is the industry standard for Python data cleaning because it offers powerful built-in functions for handling missing data, removing duplicates, data type conversion, and filtering outliers—all with simple, readable syntax that saves hours of manual coding.
Ready to Master Pandas Data Cleaning?
Learn advanced techniques for handling missing values, duplicates, and outliers in the next section.
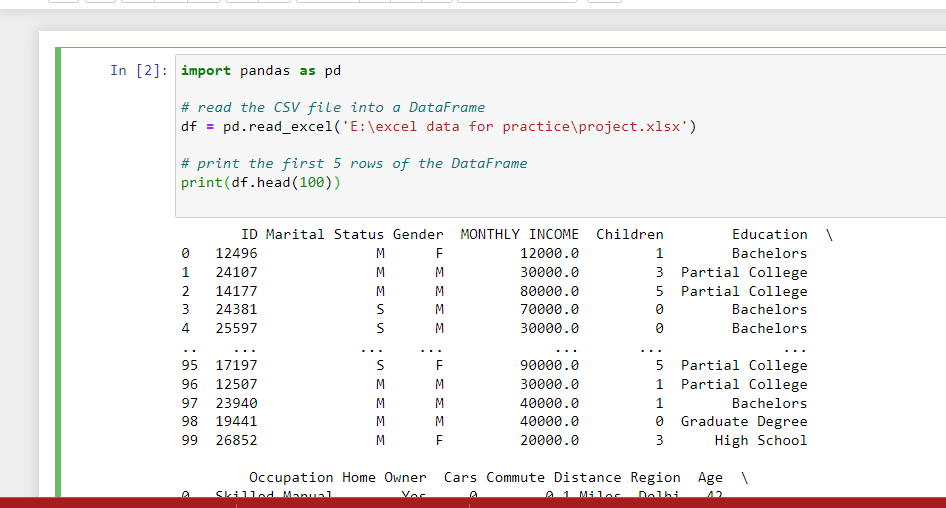
Importing CSV & Excel Files in Pandas
A common step in data cleaning with Python is importing data from
external files. With Pandas, you can easily load CSV and Excel files
into a DataFrame (df) for further analysis. This is where terms like
“csv to pandas” or “python import csv as dataframe” come in.
1. Import a CSV File into Pandas
Answer: Use the read_csv() function to load CSV data
directly into a DataFrame.
import pandas as pd
# Load CSV into DataFrame
df = pd.read_csv("data.csv")
# Preview first 5 rows
print(df.head())
👉 This is the most common way for pandas load csv and import csv python pandas.
2. Import an Excel File into Pandas
Answer: Use the read_excel() function to load Excel
sheets into Pandas.
# Load Excel file
df_excel = pd.read_excel("data.xlsx")
# Display first 5 rows
print(df_excel.head())
3. Quick Data Inspection
Answer: After importing, use
head() to preview rows and describe()
to get statistical summaries.
# Preview first 10 rows
print(df.head(10))
# Get data summary
print(df.describe())
📊 Now that you can load files into Pandas, let’s move to data cleaning methods such as handling missing values.
Next: Handling Missing Values →
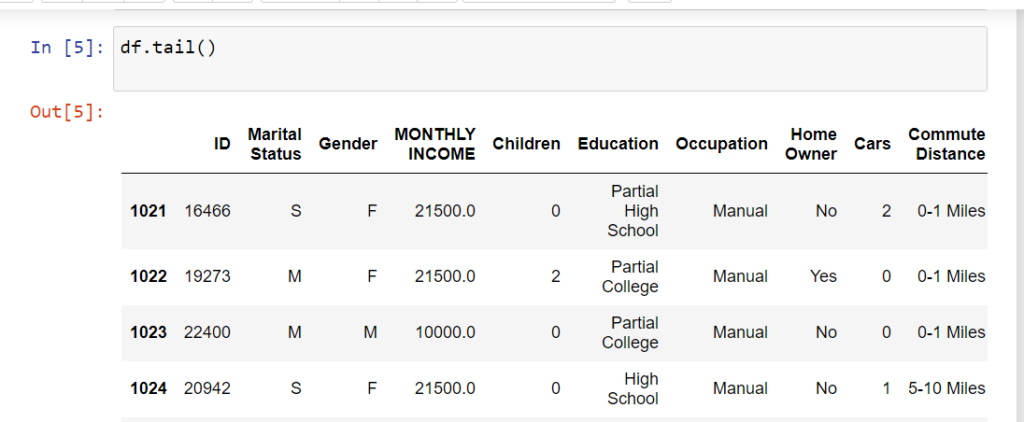
Pandas head() Method in Python
The head() function in Pandas is used to quickly preview the first few rows of a DataFrame (df).
Commonly searched as “pandas head method” or “what is df in Python”.
import pandas as pd
# Sample DataFrame
data = {'Name': ['Amit', 'Priya', 'Ravi', 'Sneha', 'Karan', 'Anjali'],
'Age': [23, 25, 22, 24, 28, 21]}
df = pd.DataFrame(data)
# Display first 5 rows
print(df.head())
# Display first 3 rows
print(df.head(3))
Name Age
0 Amit 23
1 Priya 25
2 Ravi 22
3 Sneha 24
4 Karan 28
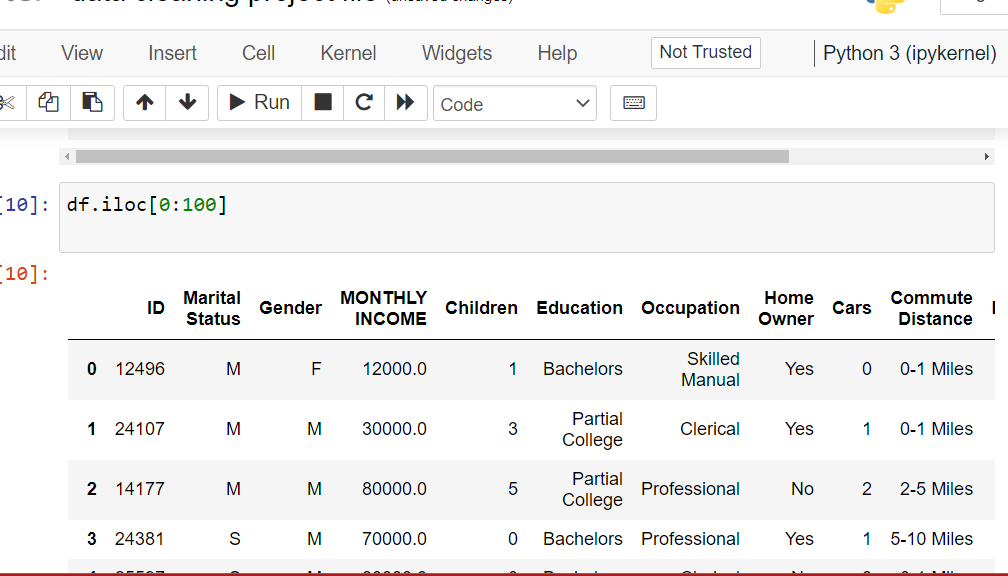
Pandas iloc – Integer Indexing
Select rows and columns by position
Use iloc to select rows and columns by integer index.
This is one of the most essential techniques for data cleaning in Python
and manipulating df in Python. Understanding iloc helps you
filter specific rows, extract subsets, and perform targeted data operations.
Code Example: Using iloc for DataFrame Indexing
Let’s create a sample DataFrame and explore different ways to use iloc:
import pandas as pd
# Sample DataFrame
data = {'Name': ['Amit', 'Priya', 'Ravi', 'Sneha', 'Karan'],
'Age': [23, 25, 22, 24, 28]}
df = pd.DataFrame(data)
# Select single row (first row)
print(df.iloc[0])
# Select multiple rows (first 3 rows)
print(df.iloc[0:3])
# Select specific cell (row 1, column 'Name')
print(df.iloc[1, 0])
Output:
Name Amit
Age 23
Name: 0, dtype: object
Name Age
0 Amit 23
1 Priya 25
2 Ravi 22
Priya
Understanding iloc Syntax
Single Row Selection
df.iloc[0] returns the first row as a Series object with column names as index.
Multiple Rows
df.iloc[0:3] selects rows from index 0 to 2 (3 is excluded). Returns a DataFrame.
Specific Cell
df.iloc[1, 0] gets the value at row 1, column 0. First number = row, second = column.
More iloc Examples for Data Cleaning
Select Last N Rows
# Get last 3 rows print(df.iloc[-3:]) # Get last row only print(df.iloc[-1])
Use negative indexing to select from the end of the DataFrame.
Select Specific Columns by Position
# Get all rows, first column only print(df.iloc[:, 0]) # Get all rows, multiple columns (0 and 1) print(df.iloc[:, [0, 1]])
The colon : means “all rows”, then specify column positions.
Combine Row and Column Selection
# Get first 3 rows, all columns print(df.iloc[0:3, :]) # Get first 2 rows, first column print(df.iloc[0:2, 0]) # Get specific rows and columns print(df.iloc[[0, 2, 4], [0, 1]])
Use lists inside iloc to select non-consecutive rows or columns.
When to Use iloc in Data Cleaning
Remove First/Last Rows
Clean headers or footers: df = df.iloc[1:-1] removes first and last rows.
Sample Data Preview
Quick inspection: df.iloc[:10] shows first 10 rows without using head().
Extract Training Data
Split datasets: train = df.iloc[:800] and test = df.iloc[800:] for ML models.
💡 Pro Tips for iloc
- Zero-based indexing: First row is 0, not 1
- iloc vs loc:
ilocuses positions (integers),locuses labels (names) - Performance:
ilocis faster for large datasets when you know exact positions - Slicing:
[start:end]excludes the end index (like Python lists)
Master DataFrame Manipulation
Next up: Learn loc for label-based indexing and conditional filtering in Pandas.
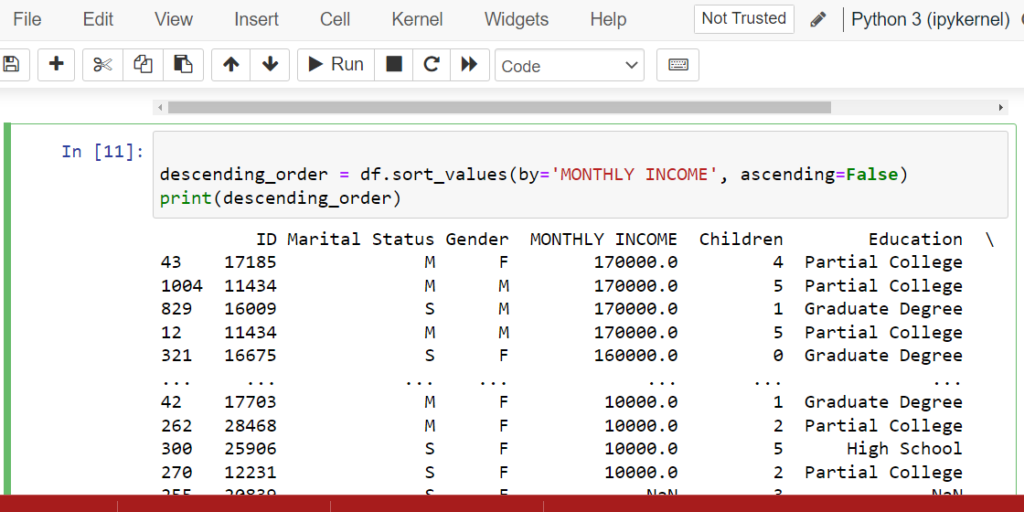
Sorting Data in Pandas (sort_values() & sort_index())
Organize your data for better analysis
Use sort_values() to sort by column values and sort_index() to sort by index.
Sorting is essential for data cleaning in Python when you need to identify top/bottom values,
detect patterns, or prepare data for visualization. Master these pandas sort methods to
streamline your data analysis workflow.
Basic Sorting Examples
Let’s create a DataFrame and explore both sorting methods:
import pandas as pd
# Sample DataFrame
data = {'Name': ['Amit', 'Priya', 'Ravi', 'Sneha'],
'Monthly Income': [50000, 70000, 45000, 60000]}
df = pd.DataFrame(data)
# Sort by index (descending)
print(df.sort_index(axis=0, ascending=False))
# Sort by column values (Monthly Income descending)
descending_order = df.sort_values(by='Monthly Income', ascending=False)
print(descending_order)
Output:
Name Monthly Income
3 Sneha 60000
2 Ravi 45000
1 Priya 70000
0 Amit 50000
Name Monthly Income
1 Priya 70000
3 Sneha 60000
0 Amit 50000
2 Ravi 45000
Understanding Pandas Sorting Methods
sort_values()
Sorts DataFrame by one or more column values. Use by='column_name' parameter. Default is ascending order.
sort_index()
Sorts DataFrame by row index or column names. Use axis=0 for rows, axis=1 for columns.
ascending Parameter
Set ascending=False for descending order (highest to lowest). Works with both methods.
Advanced Sorting Techniques
Sort by Multiple Columns
# Sort by Age (ascending), then by Salary (descending)
df_sorted = df.sort_values(by=['Age', 'Salary'],
ascending=[True, False])
print(df_sorted)
Pass a list of column names to by parameter. Use list of booleans for different sort orders.
In-Place Sorting
# Modify original DataFrame instead of creating new one df.sort_values(by='Monthly Income', inplace=True) print(df)
Use inplace=True to sort the DataFrame permanently without reassignment.
Handle Missing Values (NaN)
# Put NaN values at the beginning df.sort_values(by='Age', na_position='first') # Put NaN values at the end (default) df.sort_values(by='Age', na_position='last')
Control where NaN values appear using na_position parameter.
Reset Index After Sorting
# Sort and reset index to sequential numbers df_sorted = df.sort_values(by='Name').reset_index(drop=True) print(df_sorted)
Use reset_index(drop=True) to get clean sequential index after sorting.
Real-World Sorting Use Cases
Find Top Performers
top_5 = df.sort_values('Sales', ascending=False).head(5) – Get top 5 sales records instantly.
Identify Outliers
lowest = df.sort_values('Price').head(10) – Spot unusually low prices for data validation.
Alphabetical Ordering
df.sort_values('Name') – Sort customer names, product lists, or categories alphabetically.
Time Series Analysis
df.sort_values('Date') – Arrange data chronologically for trend analysis and forecasting.
Understanding “Ascending” in Jupyter Notebooks
Many users search for “jupyter ascending” when learning Pandas sorting.
The ascending parameter works the same in Jupyter notebooks as in regular Python scripts:
ascending=True (Default)
Smallest to largest (A to Z, 1 to 100). Perfect for finding minimum values or starting alphabetically.
ascending=False
Largest to smallest (Z to A, 100 to 1). Use for rankings, top performers, or reverse alphabetical order.
💡 Sorting Best Practices
- Performance:
sort_values()creates a new DataFrame by default – useinplace=Truefor large datasets - Stability: Pandas uses stable sorting – original order preserved for equal values
- String sorting: Case-sensitive by default – use
key=lambda x: x.str.lower()for case-insensitive - Index sorting: Use
sort_index()after operations that mess up row order - Multiple columns: Earlier columns in the list take priority in sorting
Quick Reference: Common Sorting Patterns
Sort by single column (ascending)
df.sort_values('Age')
Sort by single column (descending)
df.sort_values('Salary', ascending=False)
Sort by multiple columns
df.sort_values(['City', 'Age'])
Sort index in reverse
df.sort_index(ascending=False)
Sort and keep top 10
df.sort_values('Score', ascending=False).head(10)
Sort columns alphabetically
df.sort_index(axis=1)
Continue Learning Data Cleaning
Next: Discover how to filter and select data using conditions and boolean indexing in Pandas.
Handling Missing Values: fillna() & dropna()
Fill or remove missing data in your DataFrame
After detecting missing values with isnull(), the next step is handling them. Use fillna()
to replace missing values with specific data, or dropna() to remove rows/columns with missing data.
These are essential data cleaning techniques in Python for preparing datasets for analysis and
machine learning models.
Using fillna() to Replace Missing Values
The fillna() method fills missing values with a specified value or strategy:
import pandas as pd
import numpy as np
# Sample DataFrame with missing values
data = {'Name': ['Amit', 'Priya', 'Ravi', 'Sneha'],
'Age': [25, np.nan, 30, np.nan],
'Salary': [50000, 60000, np.nan, 70000]}
df = pd.DataFrame(data)
print("Original DataFrame:")
print(df)
# Fill missing values with 0
df_filled = df.fillna(0)
print("\nAfter fillna(0):")
print(df_filled)
# Fill with column mean
df['Age'].fillna(df['Age'].mean(), inplace=True)
print("\nAfter filling Age with mean:")
print(df)
Output:
Original DataFrame:
Name Age Salary
0 Amit 25.0 50000.0
1 Priya NaN 60000.0
2 Ravi 30.0 NaN
3 Sneha NaN 70000.0
After fillna(0):
Name Age Salary
0 Amit 25.0 50000.0
1 Priya 0.0 60000.0
2 Ravi 30.0 0.0
3 Sneha 0.0 70000.0
After filling Age with mean:
Name Age Salary
0 Amit 25.0 50000.0
1 Priya 27.5 60000.0
2 Ravi 30.0 NaN
3 Sneha 27.5 70000.0
Common fillna() Strategies
Fill with Constant
df.fillna(0) or df.fillna('Unknown') – Replace all NaN with a fixed value.
Fill with Mean/Median
df.fillna(df.mean()) – Use statistical measures for numerical columns. Best for normally distributed data.
Forward Fill (ffill)
df.fillna(method='ffill') – Copy previous value downward. Great for time series data.
Backward Fill (bfill)
df.fillna(method='bfill') – Copy next value upward. Useful when future data is more relevant.
Advanced fillna() Techniques
Fill Different Columns with Different Values
# Fill each column with specific value
fill_values = {'Age': 0, 'Salary': df['Salary'].median(), 'City': 'Unknown'}
df.fillna(value=fill_values, inplace=True)
print(df)
Use a dictionary to specify different fill values for each column.
Fill with Column Mode (Most Frequent Value)
# Fill categorical columns with mode
df['Category'].fillna(df['Category'].mode()[0], inplace=True)
# Or for multiple columns
for col in ['City', 'Department']:
df[col].fillna(df[col].mode()[0], inplace=True)
Best for categorical data – fills with the most common value.
Linear Interpolation for Time Series
# Interpolate missing values df['Temperature'].interpolate(method='linear', inplace=True) # For time-based interpolation df['Sales'].interpolate(method='time', inplace=True)
Estimates missing values based on surrounding data points. Perfect for continuous data.
Conditional Filling Based on Other Columns
# Fill based on condition
df.loc[df['City'] == 'Delhi', 'Salary'] = \
df.loc[df['City'] == 'Delhi', 'Salary'].fillna(75000)
# Or using np.where
import numpy as np
df['Age'] = np.where(df['Age'].isnull(),
df.groupby('Department')['Age'].transform('mean'),
df['Age'])
Fill missing values differently based on category or group.
Using dropna() to Remove Missing Values
The dropna() method removes rows or columns containing missing values:
import pandas as pd
import numpy as np
# Sample DataFrame
data = {'A': [1, 2, np.nan, 4],
'B': [5, np.nan, np.nan, 8],
'C': [9, 10, 11, 12]}
df = pd.DataFrame(data)
print("Original DataFrame:")
print(df)
# Drop rows with any missing values
df_dropped = df.dropna()
print("\nAfter dropna() - Remove rows:")
print(df_dropped)
# Drop columns with any missing values
df_dropped_cols = df.dropna(axis=1)
print("\nAfter dropna(axis=1) - Remove columns:")
print(df_dropped_cols)
Output:
Original DataFrame:
A B C
0 1.0 5.0 9
1 2.0 NaN 10
2 NaN NaN 11
3 4.0 8.0 12
After dropna() - Remove rows:
A B C
0 1.0 5.0 9
3 4.0 8.0 12
After dropna(axis=1) - Remove columns:
C
0 9
1 10
2 11
3 12
dropna() Parameters Explained
axis Parameter
axis=0 (default) drops rows, axis=1 drops columns with missing values.
how Parameter
how='any' (default) drops if ANY value is missing, how='all' only if ALL values missing.
thresh Parameter
thresh=3 keeps rows with at least 3 non-null values. Good for sparse datasets.
subset Parameter
subset=['Age', 'Salary'] only considers specific columns when dropping.
Advanced dropna() Examples
Drop Rows with All Missing Values
# Only drop completely empty rows
df.dropna(how='all', inplace=True)
# Example use case
df = pd.DataFrame({
'A': [1, np.nan, np.nan],
'B': [2, np.nan, np.nan],
'C': [3, 4, np.nan]
})
df.dropna(how='all') # Only row 1 is dropped
Useful when you have placeholder rows that are completely empty.
Keep Rows with Minimum Non-Null Values
# Keep rows with at least 3 non-null values df.dropna(thresh=3, inplace=True) # For a 5-column dataset, keep rows with at least 80% data min_count = int(len(df.columns) * 0.8) df.dropna(thresh=min_count, inplace=True)
Balance between keeping data and maintaining quality.
Drop Based on Specific Columns Only
# Only drop if critical columns have missing values df.dropna(subset=['CustomerID', 'OrderDate'], inplace=True) # Keep rows where at least one key column is filled df.dropna(subset=['Email', 'Phone'], how='all', inplace=True)
Preserve data by only checking columns that are absolutely required.
Drop Columns with High Missing Percentage
# Drop columns with more than 50% missing values
threshold = 0.5
missing_pct = df.isnull().mean()
cols_to_drop = missing_pct[missing_pct > threshold].index
df.drop(columns=cols_to_drop, inplace=True)
print(f"Dropped columns: {list(cols_to_drop)}")
Automatically remove low-quality columns from your dataset.
When to Use fillna() vs dropna()?
Use fillna() When:
- ✓ You have few missing values (less than 5%)
- ✓ Dropping would lose too much data
- ✓ Missing data follows a pattern (time series)
- ✓ You can estimate reasonable replacement values
Use dropna() When:
- ✓ You have abundant data (can afford to lose rows)
- ✓ Missing values are random and unpredictable
- ✓ Critical columns have missing data
- ✓ Imputation might introduce bias
💡 Missing Value Handling Best Practices
- Document decisions: Always note why you chose to fill or drop, and what values you used
- Create copies: Use
df_clean = df.dropna()instead ofinplace=Trueuntil certain - Check impact: Compare statistics before/after –
df.describe()vsdf_clean.describe() - Domain knowledge: Consult subject matter experts before filling with mean/median
- Test both approaches: Try fillna() and dropna() separately, compare model performance
- Hybrid approach: Fill some columns, drop others based on importance and missing %
- Preserve original: Keep a backup of raw data before any transformations
Real-World Example: Complete Workflow
import pandas as pd
import numpy as np
# Load dataset
df = pd.read_csv('customer_data.csv')
# Step 1: Analyze missing values
print("Missing values per column:")
print(df.isnull().sum())
# Step 2: Handle numerical columns - fill with median
numerical_cols = ['Age', 'Income', 'Transaction_Amount']
for col in numerical_cols:
if df[col].isnull().sum() > 0:
median_value = df[col].median()
df[col].fillna(median_value, inplace=True)
print(f"Filled {col} with median: {median_value}")
# Step 3: Handle categorical columns - fill with mode
categorical_cols = ['City', 'Product_Category']
for col in categorical_cols:
if df[col].isnull().sum() > 0:
mode_value = df[col].mode()[0]
df[col].fillna(mode_value, inplace=True)
print(f"Filled {col} with mode: {mode_value}")
# Step 4: Drop rows where critical columns are missing
critical_cols = ['Customer_ID', 'Order_Date']
df.dropna(subset=critical_cols, inplace=True)
# Step 5: Drop columns with >70% missing data
threshold = 0.7
missing_pct = df.isnull().mean()
cols_to_drop = missing_pct[missing_pct > threshold].index
if len(cols_to_drop) > 0:
df.drop(columns=cols_to_drop, inplace=True)
print(f"Dropped high-missing columns: {list(cols_to_drop)}")
# Step 6: Verify cleaning
print(f"\nFinal missing values:")
print(df.isnull().sum())
print(f"\nDataset shape: {df.shape}")
# Save cleaned data
df.to_csv('customer_data_cleaned.csv', index=False)
Quick Reference: fillna() & dropna()
Fill all NaN with 0
df.fillna(0)
Fill with column mean
df['Age'].fillna(df['Age'].mean())
Forward fill (copy previous value)
df.fillna(method='ffill')
Drop rows with any missing values
df.dropna()
Drop columns with any missing values
df.dropna(axis=1)
Drop rows only if all values are NaN
df.dropna(how='all')
Keep rows with at least 3 values
df.dropna(thresh=3)
Drop based on specific columns
df.dropna(subset=['Age', 'Salary'])
Data Cleaned Successfully!
Next: Learn how to remove duplicate rows and handle data type conversions in Pandas.
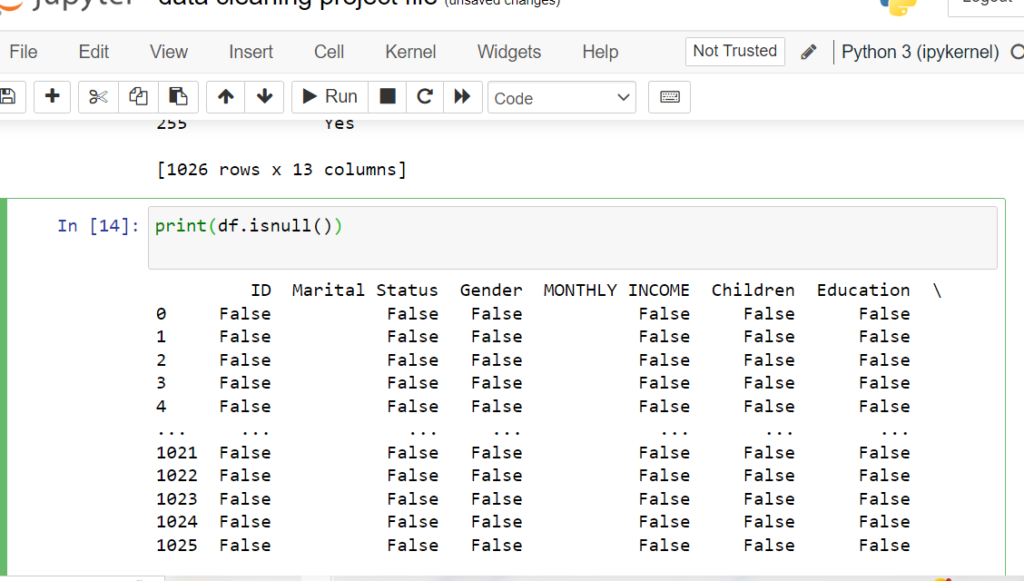
Identifying Columns & Rows with Missing Values
Pinpoint exactly where missing data exists
Use isnull() with any() and sum() to find missing values in columns,
and filter rows containing null values. This targeted approach to Python data cleansing helps
you understand the scope of missing data and make informed decisions about which columns with missing
values in Pandas need attention during data cleaning.
Finding Columns and Rows with Missing Data
Let’s identify which columns and rows contain missing values:
import pandas as pd
# Sample DataFrame
data = {'Name': ['Amit', 'Priya', 'Ravi', 'Sneha'],
'Age': [25, None, 30, None],
'City': ['Delhi', 'Mumbai', None, 'Pune']}
df = pd.DataFrame(data)
# Columns with missing values
print(df.isnull().any(axis=0))
# Count missing values per column
print(df.isnull().sum())
# Rows with missing values
missing_rows = df[df.isnull().any(axis=1)]
print(missing_rows)
Output:
Name False
Age True
City True
dtype: bool
Name 0
Age 2
City 1
dtype: int64
Name Age City
1 Priya NaN Mumbai
2 Ravi 30.0 NaN
3 Sneha NaN Pune
Understanding any() vs all()
any(axis=0)
Returns True for columns that have at least one missing value. Default behavior for column-wise check.
any(axis=1)
Returns True for rows that have at least one missing value. Use to filter incomplete rows.
all()
Returns True only if all values are missing. Useful for finding completely empty columns/rows.
Advanced Column Detection Techniques
Get List of Column Names with Missing Values
# Method 1: Using any()
cols_with_missing = df.columns[df.isnull().any()].tolist()
print(f"Columns with missing values: {cols_with_missing}")
# Method 2: Using sum() and filtering
missing_counts = df.isnull().sum()
cols_with_missing = missing_counts[missing_counts > 0].index.tolist()
print(f"Columns with missing data: {cols_with_missing}")
Returns a clean list of column names for further processing.
Find Columns with No Missing Values
# Get columns that are completely filled
complete_cols = df.columns[df.notnull().all()].tolist()
print(f"Complete columns: {complete_cols}")
# Alternative method
complete_cols = df.columns[df.isnull().sum() == 0].tolist()
print(f"Columns with no missing data: {complete_cols}")
Identify which columns are already clean and ready for analysis.
Rank Columns by Missing Value Count
# Sort columns by number of missing values (descending)
missing_counts = df.isnull().sum().sort_values(ascending=False)
print("Columns ranked by missing data:")
print(missing_counts)
# Get top 5 columns with most missing values
top_missing = df.isnull().sum().nlargest(5)
print("\nTop 5 columns with most missing values:")
print(top_missing)
Prioritize which columns need immediate attention during data cleaning.
Find Columns Above Missing Value Threshold
# Find columns with more than 30% missing values
threshold = 0.3
missing_pct = df.isnull().mean()
high_missing_cols = missing_pct[missing_pct > threshold].index.tolist()
print(f"Columns with >{threshold*100}% missing:")
for col in high_missing_cols:
pct = missing_pct[col] * 100
print(f" {col}: {pct:.2f}%")
Automatically identify low-quality columns that may need to be dropped.
Advanced Row Detection Techniques
Get Row Indices with Missing Values
# Get index positions of rows with missing data
rows_with_missing_idx = df[df.isnull().any(axis=1)].index.tolist()
print(f"Row indices with missing values: {rows_with_missing_idx}")
# Count how many rows have missing data
num_rows_missing = df.isnull().any(axis=1).sum()
print(f"Total rows with missing data: {num_rows_missing}")
Useful for logging or targeted row-by-row processing.
Find Rows with Multiple Missing Values
# Count missing values per row
missing_per_row = df.isnull().sum(axis=1)
print("Missing values per row:")
print(missing_per_row)
# Get rows with 2 or more missing values
rows_multiple_missing = df[missing_per_row >= 2]
print("\nRows with 2+ missing values:")
print(rows_multiple_missing)
Identify rows with severe data quality issues.
Find Completely Empty Rows
# Find rows where ALL values are missing
completely_empty_rows = df[df.isnull().all(axis=1)]
print("Completely empty rows:")
print(completely_empty_rows)
# Get count
num_empty = df.isnull().all(axis=1).sum()
print(f"\nNumber of completely empty rows: {num_empty}")
Detect placeholder or corrupted rows that should be removed.
Filter Rows Based on Specific Columns
# Get rows where specific columns have missing values
rows_missing_age = df[df['Age'].isnull()]
print("Rows with missing Age:")
print(rows_missing_age)
# Rows missing EITHER Age OR City
rows_missing_either = df[df[['Age', 'City']].isnull().any(axis=1)]
print("\nRows missing Age or City:")
print(rows_missing_either)
# Rows missing BOTH Age AND City
rows_missing_both = df[df[['Age', 'City']].isnull().all(axis=1)]
print("\nRows missing both Age and City:")
print(rows_missing_both)
Target specific column combinations for conditional processing.
Real-World Detection Scenarios
Data Quality Report
Create automated reports showing which columns/rows need cleaning before analysis starts.
Conditional Processing
Process different columns differently based on their missing data patterns and percentages.
Validation Checks
Ensure critical columns (IDs, dates) have no missing values before proceeding with ETL pipeline.
User Notifications
Alert users which fields are incomplete in data entry forms or CSV uploads.
Complete Missing Data Detection Workflow
import pandas as pd
import numpy as np
def comprehensive_missing_analysis(df):
"""
Complete analysis of missing values in DataFrame
"""
print("=" * 60)
print("COMPREHENSIVE MISSING VALUE DETECTION")
print("=" * 60)
# Overall statistics
total_cells = df.shape[0] * df.shape[1]
total_missing = df.isnull().sum().sum()
pct_missing = (total_missing / total_cells) * 100
print(f"\n📊 OVERALL STATISTICS")
print(f"Total cells: {total_cells:,}")
print(f"Total missing: {total_missing:,}")
print(f"Percentage missing: {pct_missing:.2f}%")
# Column analysis
print(f"\n📋 COLUMN ANALYSIS")
cols_with_missing = df.columns[df.isnull().any()].tolist()
cols_complete = df.columns[df.notnull().all()].tolist()
print(f"Columns with missing values: {len(cols_with_missing)}")
print(f"Complete columns: {len(cols_complete)}")
if cols_with_missing:
print("\nMissing value breakdown by column:")
missing_summary = df.isnull().sum()
missing_summary = missing_summary[missing_summary > 0].sort_values(ascending=False)
for col, count in missing_summary.items():
pct = (count / len(df)) * 100
print(f" • {col}: {count} ({pct:.2f}%)")
# Row analysis
print(f"\n📝 ROW ANALYSIS")
rows_with_missing = df.isnull().any(axis=1).sum()
rows_complete = df.notnull().all(axis=1).sum()
completely_empty = df.isnull().all(axis=1).sum()
print(f"Rows with missing values: {rows_with_missing} ({(rows_with_missing/len(df)*100):.2f}%)")
print(f"Complete rows: {rows_complete} ({(rows_complete/len(df)*100):.2f}%)")
print(f"Completely empty rows: {completely_empty}")
# Missing value distribution
missing_per_row = df.isnull().sum(axis=1)
print(f"\nMissing value distribution per row:")
print(missing_per_row.value_counts().sort_index())
# Critical columns check (example)
critical_cols = ['Customer_ID', 'Order_Date', 'Amount']
existing_critical = [col for col in critical_cols if col in df.columns]
if existing_critical:
print(f"\n⚠️ CRITICAL COLUMNS CHECK")
for col in existing_critical:
missing = df[col].isnull().sum()
if missing > 0:
print(f" ⚠️ WARNING: {col} has {missing} missing values!")
else:
print(f" ✓ {col} is complete")
return {
'columns_with_missing': cols_with_missing,
'rows_with_missing': rows_with_missing,
'total_missing': total_missing
}
# Usage
results = comprehensive_missing_analysis(df)
print(f"\nColumns needing attention: {results['columns_with_missing']}")
💡 Detection Best Practices
- Always detect first: Run detection analysis before any filling or dropping operations
- Set thresholds: Define acceptable missing % for your use case (e.g., 5% warning, 30% critical)
- Document patterns: Note if missing values are random or follow patterns (e.g., all weekend dates missing)
- Check critical columns: Verify ID, timestamp, and key business columns have no missing values
- Visualize distribution: Use
df.isnull().sum().plot(kind='bar')for quick overview - Cross-check: Missing values in related columns often indicate systematic issues
Quick Reference: Finding Missing Values
Check if any column has missing values
df.isnull().any(axis=0)
Get column names with missing values
df.columns[df.isnull().any()].tolist()
Count missing values per column
df.isnull().sum()
Filter rows with any missing values
df[df.isnull().any(axis=1)]
Get rows with complete data only
df[df.notnull().all(axis=1)]
Count rows with missing values
df.isnull().any(axis=1).sum()
Find completely empty rows
df[df.isnull().all(axis=1)]
Missing values in specific column
df[df['ColumnName'].isnull()]
Master Data Cleaning Detection
Next: Learn how to handle duplicate rows and ensure data uniqueness in your DataFrames.
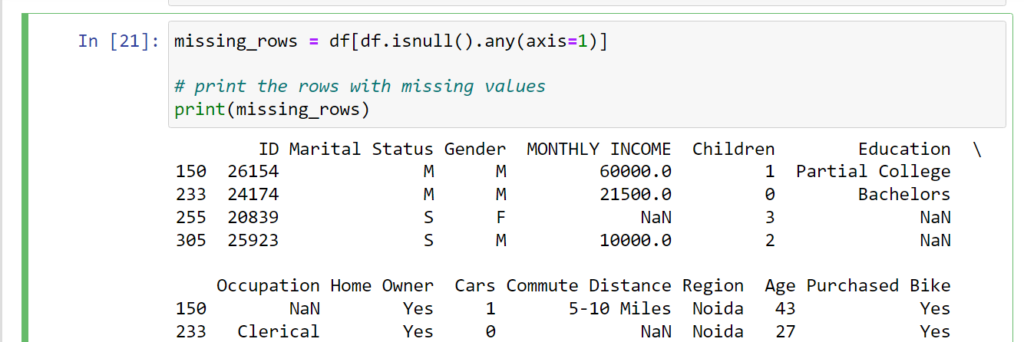
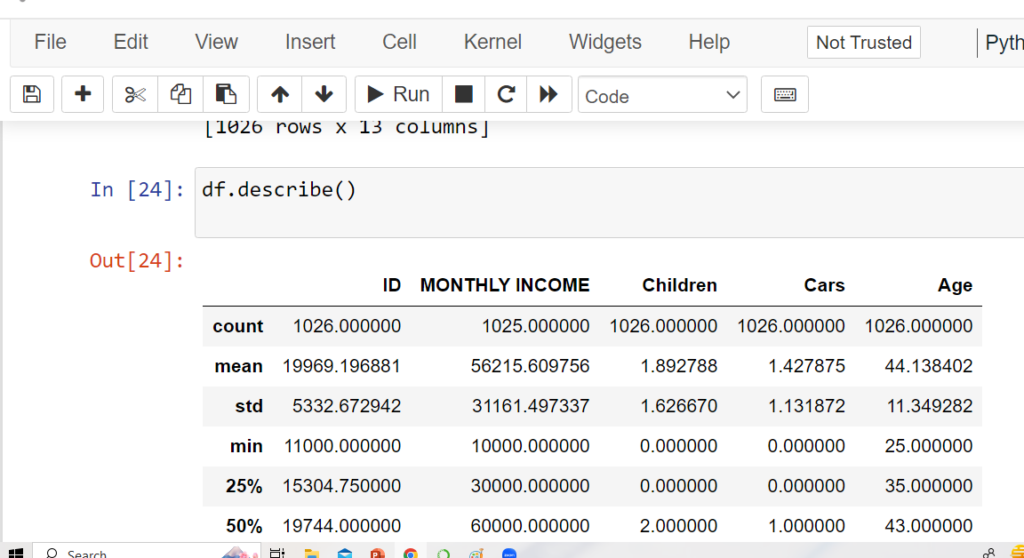
Descriptive Statistics with describe()
The describe() method summarizes numerical columns: count, mean, std, min,
25%, 50% (median), 75%, and max values.
Keywords: “pandas describe method”, “data cleaning python pandas”.
import pandas as pd
# Sample DataFrame
data = {'Age': [25, 30, 35, 40, 28],
'Income': [40000, 50000, 60000, 75000, 48000]}
df = pd.DataFrame(data)
# Descriptive statistics
print(df.describe())
Age Income
count 5.000000 5.000000
mean 31.600000 54600.000000
std 6.429101 14031.787366
min 25.000000 40000.000000
25% 28.000000 48000.000000
50% 30.000000 50000.000000
75% 35.000000 60000.000000
max 40.000000 75000.000000
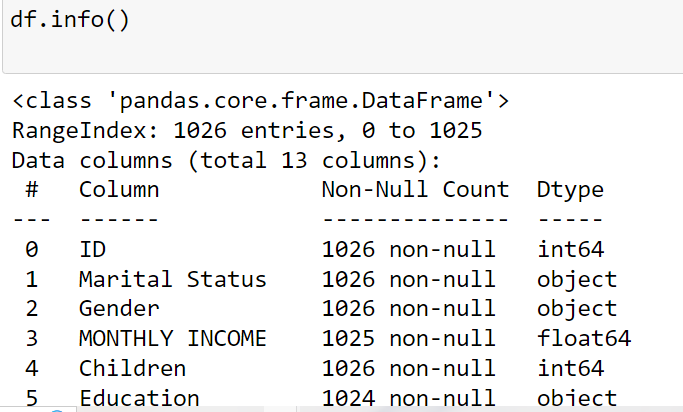
info()
DataFrame Summary with info()
The info() method displays column names, data types, non-null counts,
and memory usage of a DataFrame.
Keywords: “df in python”, “what is df in python”, “python data cleansing”.
import pandas as pd
# Sample DataFrame
data = {'Name': ['Amit', 'Priya', 'Ravi'],
'Age': [25, 30, None],
'Country': ['India', 'USA', 'UK']}
df = pd.DataFrame(data)
# Get DataFrame info
df.info()
RangeIndex: 3 entries, 0 to 2 Data columns (total 3 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 Name 3 non-null object 1 Age 2 non-null float64 2 Country 3 non-null object dtypes: float64(1), object(2) memory usage: 200.0+ bytes
Handling Missing Data with fillna()
Use fillna() to replace NaN values in different ways: constant values,
forward/backward fill, interpolation, mean/median, or custom logic.
Keywords: “fill missing values with median pandas”, “python pandas data cleaning”.
# Fill missing values with constant
df.fillna(0)
# Fill with previous value
df.fillna(method='ffill')
# Fill with interpolated values
df.interpolate()
# Fill missing values with mean
df.fillna(value=df.mean())
# Fill missing values with median
df.fillna(value=df.median())
# Fill using custom function
def my_func():
return 99
df.fillna(value=my_func())
# Fill specific column
df['Education'] = df['Education'].fillna(value='Partial College', inplace=False)
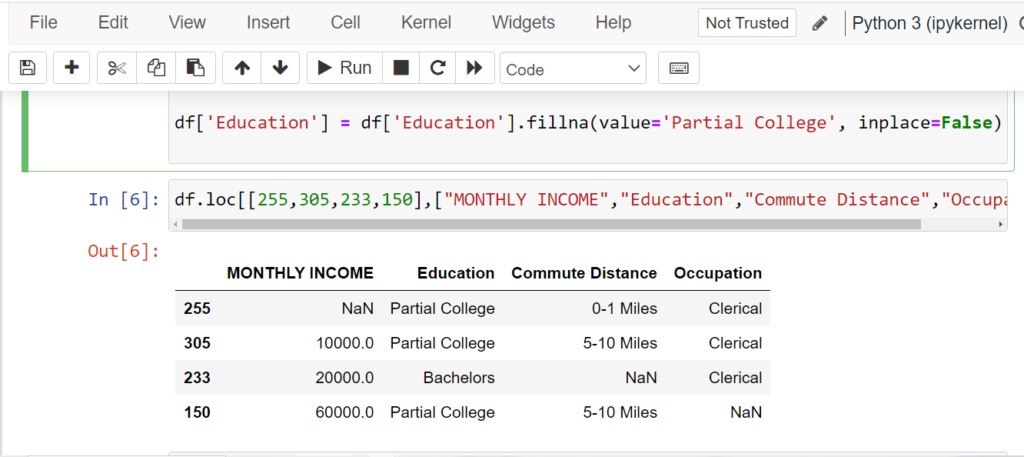
Removing Missing Data with dropna()
Use dropna() to remove rows or columns with missing values in a DataFrame.
Keywords: “pandas data cleaning methods”, “data cleaning python pandas”.
# Drop rows with any NaN values
df.dropna()
# Drop rows where all values are NaN
df.dropna(how='all')
# Drop columns with NaN values
df.dropna(axis=1)
# Keep rows with at least 2 non-NaN values
df.dropna(thresh=2)
# Drop rows where 'Age' is NaN
df.dropna(subset=['Age'])
Replacing Values with replace()
The replace() method is used to substitute specific values in a DataFrame or Series.
Keywords: “pandas data cleaning methods”, “python data cleansing”.
# Replace value 0 with NaN
import numpy as np
df.replace(0, np.nan)
# Replace multiple values
df.replace(['?', 'NA', 'missing'], np.nan)
# Replace using column-specific mapping
df.replace({'Gender': {'M': 'Male', 'F': 'Female'}})
# Replace text patterns using regex
df.replace(to_replace=r'^Unknown.*', value='Other', regex=True)
# Replace 0 in 'Income' column with column median
df['Income'].replace(0, df['Income'].median(), inplace=True)
Handling Duplicates with duplicated() and drop_duplicates()
Use duplicated() to detect duplicate rows and drop_duplicates() to remove them.
Keywords: “pandas data cleaning methods”, “python pandas data cleaning”.
# Find duplicate rows
df.duplicated()
# Count duplicate rows
df.duplicated().sum()
# Drop duplicate rows
df.drop_duplicates()
# Drop duplicates based on 'Name' column
df.drop_duplicates(subset=['Name'])
# Keep last occurrence, drop others
df.drop_duplicates(keep='last')
Custom Cleaning with apply() and Lambda Functions
The apply() method lets you apply a custom function (or lambda)
to each element, row, or column in a DataFrame.
Keywords: “python data cleansing”, “data cleaning python pandas”.
# Convert all names to lowercase
df['Name'] = df['Name'].apply(lambda x: x.lower())
# Remove leading/trailing spaces
df['City'] = df['City'].apply(lambda x: x.strip())
# Replace negative values with 0
df['Income'] = df['Income'].apply(lambda x: max(x, 0))
# Create full_name by combining two columns
df['Full_Name'] = df.apply(lambda row: row['First'] + ' ' + row['Last'], axis=1)
# Define a custom function
def clean_age(x):
return 0 if pd.isnull(x) else int(x)
df['Age'] = df['Age'].apply(clean_age)
Data cleaning, also known as data cleansing or data scrubbing, is the process of identifying and correcting or removing errors and inconsistencies in datasets to improve data quality and ensure it is suitable for analysis
Common data cleaning tasks include handling missing values, removing duplicates, correcting data types, standardizing formats, dealing with outliers, and handling inconsistencies in data.
Imputation is the process of filling in missing values with estimated or calculated values. You can use Pandas functions like fillna() or libraries like Scikit-Learn’s SimpleImputer for imputation.
You can handle missing values using libraries like Pandas by using functions like dropna() to remove rows with missing values, fillna() to fill missing values with a specified value, or by using techniques like interpolation or imputation.
You can remove duplicate rows in a Pandas DataFrame using the drop_duplicates() method.
Outliers are data points that deviate significantly from the rest of the data. You can detect and handle outliers using techniques such as Z-score, IQR (Interquartile Range), or visualization methods. Libraries like Scikit-Learn and Matplotlib can be helpful.